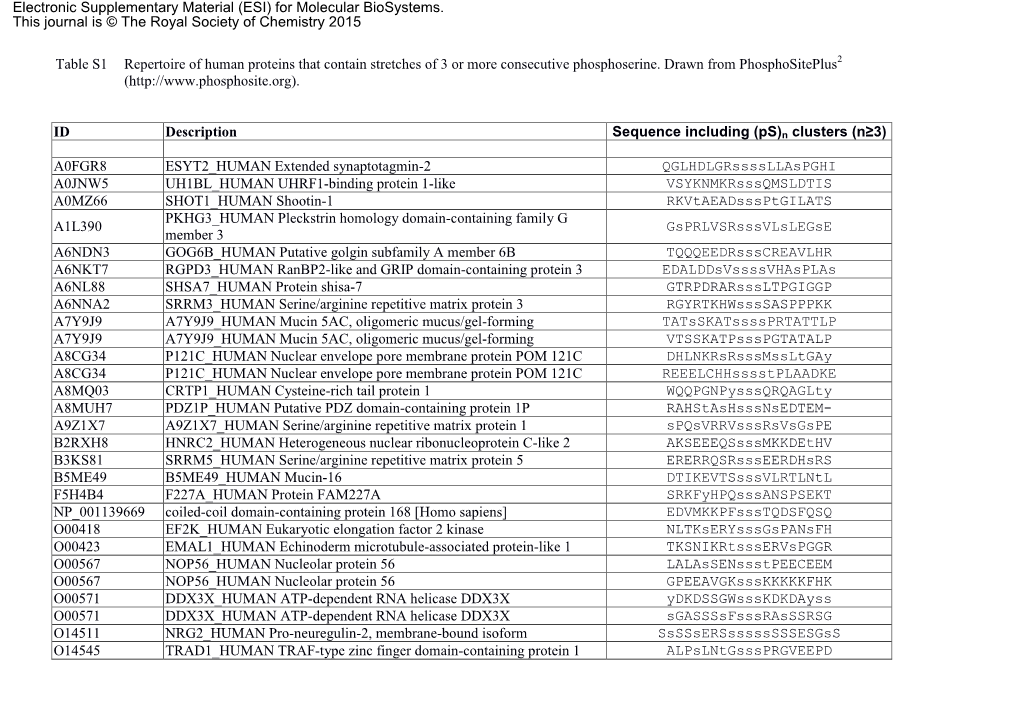
Table S1 Repertoire of Human Proteins That Contain Stretches of 3 Or More Consecutive Phosphoserine
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1
Supplementary Table 1. Phosphoryl I-Area II-Area II-Debunker III-Area Gene Symbol Protein Name Phosphorylated Peptide I-R2 I-Debunker score II-R2 III-R2 ation Site Ratio Ratio score Ratio 92154 ABBA-1 Actin-bundling protein with BAIAP2 homologyK.TPTVPDS*PGYMGPTR.A S601 1.76 0.95 0.999958250 1.18 0.98 0.999971836 0.98 0.98 92154 ABBA-1 Actin-bundling protein with BAIAP2 homologyR.AGS*EECVFYTDETASPLAPDLAK.A S612 1.40 0.99 0.999977464 1.03 0.99 0.999986373 1.00 0.99 92154 ABBA-1 Actin-bundling protein with BAIAP2 homologyK.GGGAPWPGGAQTYS*PSSTCR.Y S300 0.49 0.98 0.999985406 0.97 0.99 0.999983906 2.03 0.97 23 ABCF1 ATP-binding cassette sub-family F memberK.QQPPEPEWIGDGESTS*PSDK.V 1 S22 1.09 0.98 0.872361494 0.81 1.00 0.847115585 0.97 0.97 27 ABL2 Isoform IA of Tyrosine-protein kinase ABL2K.VPVLIS*PTLK.H S936 0.76 0.98 0.999991559 1.06 0.99 0.999989547 0.99 0.96 3983 ABLIM1 Actin binding LIM protein 1 R.TLS*PTPSAEGYQDVR.D S433 1.27 0.99 0.999994010 1.16 0.98 0.999989861 1.11 0.99 31 ACACA acetyl-Coenzyme A carboxylase alpha isoformR.FIIGSVSEDNS*EDEISNLVK.L 1 S29 1.23 0.99 0.999992898 0.72 0.99 0.999992499 0.83 0.99 65057 ACD Adrenocortical dysplasia protein homologR.TPS*SPLQSCTPSLSPR.S S424 1.51 0.90 0.999936226 0.82 0.88 0.997623142 0.46 0.93 22985 ACIN1 Apoptotic chromatin condensation inducerK.ASLVALPEQTASEEET*PPPLLTK.E in the nucleus T414 0.90 0.92 0.922696554 0.91 0.92 0.993049132 0.95 0.99 22985 ACIN1 Apoptotic chromatin condensation inducerK.ASLVALPEQTAS*EEETPPPLLTK.E in the nucleus S410 1.02 0.99 0.997470008 0.80 0.99 0.999702808 0.96 -

THE DEVELOPMENT of CHEMICAL METHODS to DISCOVER KINASE SUBSTRATES and MAP CELL SIGNALING with GAMMA-MODIFIED ATP ANALOG-DEPENDENT KINASE-CATALYZED PHOSPHORYLATION By
Wayne State University Wayne State University Dissertations 1-1-2017 The evelopmeD nt Of Chemical Methods To Discover Kinase Substrates And Map Cell Signaling With Gamma-Modified Atp Analog- Dependent Kinase-Catalyzed Phosphorylation Dissanayaka Mudiyanselage Maheeka Madhubashini Embogama Wayne State University, Follow this and additional works at: https://digitalcommons.wayne.edu/oa_dissertations Part of the Analytical Chemistry Commons, and the Biochemistry Commons Recommended Citation Embogama, Dissanayaka Mudiyanselage Maheeka Madhubashini, "The eD velopment Of Chemical Methods To Discover Kinase Substrates And Map Cell Signaling With Gamma-Modified Atp Analog-Dependent Kinase-Catalyzed Phosphorylation" (2017). Wayne State University Dissertations. 1698. https://digitalcommons.wayne.edu/oa_dissertations/1698 This Open Access Dissertation is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Dissertations by an authorized administrator of DigitalCommons@WayneState. THE DEVELOPMENT OF CHEMICAL METHODS TO DISCOVER KINASE SUBSTRATES AND MAP CELL SIGNALING WITH GAMMA-MODIFIED ATP ANALOG-DEPENDENT KINASE-CATALYZED PHOSPHORYLATION by DISSANAYAKA M. MAHEEKA M. EMBOGAMA DISSERTATION Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY 2017 MAJOR: CHEMISTRY (Biochemistry) Approved By: Advisor Date DEDICATION To my beloved mother, father, husband, daughter and sister. ii ACKNOWLEGEMENTS Many people have helped me during the past five years of earning my PhD. I would like to take this opportunity to convey my gratitude to them. First and foremost, I would like to thank my research supervisor Dr. Mary Kay Pflum for being the greatest mentor that I have met so far. -

Produktinformation
Produktinformation Diagnostik & molekulare Diagnostik Laborgeräte & Service Zellkultur & Verbrauchsmaterial Forschungsprodukte & Biochemikalien Weitere Information auf den folgenden Seiten! See the following pages for more information! Lieferung & Zahlungsart Lieferung: frei Haus Bestellung auf Rechnung SZABO-SCANDIC Lieferung: € 10,- HandelsgmbH & Co KG Erstbestellung Vorauskassa Quellenstraße 110, A-1100 Wien T. +43(0)1 489 3961-0 Zuschläge F. +43(0)1 489 3961-7 [email protected] • Mindermengenzuschlag www.szabo-scandic.com • Trockeneiszuschlag • Gefahrgutzuschlag linkedin.com/company/szaboscandic • Expressversand facebook.com/szaboscandic SANTA CRUZ BIOTECHNOLOGY, INC. FAM214A siRNA (m): sc-141565 BACKGROUND STORAGE AND RESUSPENSION FAM214A, also known as BC031353 or KIAA1370, is a 1,076 amino acid Store lyophilized siRNA duplex at -20° C with desiccant. Stable for at least protein that is alternatively spliced into two isoforms. The gene encoding one year from the date of shipment. Once resuspended, store at -20° C, KIAA1370 maps to human chromosome 15, which encodes more than avoid contact with RNAses and repeated freeze thaw cycles. 700 genes and comprises about 3% of the human genome. Angelman and Resuspend lyophilized siRNA duplex in 330 µl of the RNAse-free water Prader-Willi syndromes are associated with loss of function or deletion of provided. Resuspension of the siRNA duplex in 330 µl of RNAse-free water genes in the 15q11-q13 region. In Angelman syndrome, this loss is due to makes a 10 µM solution in a 10 µM Tris-HCl, pH 8.0, 20 mM NaCl, 1 mM inactivity of the maternal 15q21.2 encoded UBE3A gene in the brain by EDTA buffered solution. either chromosomal deletion or mutation. -

The VE-Cadherin/Amotl2 Mechanosensory Pathway Suppresses Aortic In�Ammation and the Formation of Abdominal Aortic Aneurysms
The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang Karolinska Institute Evelyn Hutterer Karolinska Institute Sara Hultin Karolinska Institute Otto Bergman Karolinska Institute Maria Forteza Karolinska Institute Zorana Andonovic Karolinska Institute Daniel Ketelhuth Karolinska University Hospital, Stockholm, Sweden Joy Roy Karolinska Institute Per Eriksson Karolinska Institute Lars Holmgren ( [email protected] ) Karolinska Institute Article Keywords: arterial endothelial cells (ECs), vascular disease, abdominal aortic aneurysms Posted Date: June 15th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-600069/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inflammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang1, Evelyn Hutterer1, Sara Hultin1, Otto Bergman2, Maria J. Forteza2, Zorana Andonovic1, Daniel F.J. Ketelhuth2,3, Joy Roy4, Per Eriksson2 and Lars Holmgren1*. 1Department of Oncology-Pathology, BioClinicum, Karolinska Institutet, Stockholm, Sweden. 2Department of Medicine Solna, BioClinicum, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden. 3Department of Cardiovascular and Renal Research, Institutet of Molecular Medicine, Univ. of Southern Denmark, Odense, Denmark 4Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital, Stockholm, -

Antibodies Products
Chapter 2 : Gentaur Products List • Human Signal peptidase complex catalytic subunit • Human Sjoegren syndrome nuclear autoantigen 1 SSNA1 • Human Small proline rich protein 2A SPRR2A ELISA kit SEC11A SEC11A ELISA kit SpeciesHuman ELISA kit SpeciesHuman SpeciesHuman • Human Signal peptidase complex catalytic subunit • Human Sjoegren syndrome scleroderma autoantigen 1 • Human Small proline rich protein 2B SPRR2B ELISA kit SEC11C SEC11C ELISA kit SpeciesHuman SSSCA1 ELISA kit SpeciesHuman SpeciesHuman • Human Signal peptidase complex subunit 1 SPCS1 ELISA • Human Ski oncogene SKI ELISA kit SpeciesHuman • Human Small proline rich protein 2D SPRR2D ELISA kit kit SpeciesHuman • Human Ski like protein SKIL ELISA kit SpeciesHuman SpeciesHuman • Human Signal peptidase complex subunit 2 SPCS2 ELISA • Human Skin specific protein 32 C1orf68 ELISA kit • Human Small proline rich protein 2E SPRR2E ELISA kit kit SpeciesHuman SpeciesHuman SpeciesHuman • Human Signal peptidase complex subunit 3 SPCS3 ELISA • Human SLAIN motif containing protein 1 SLAIN1 ELISA kit • Human Small proline rich protein 2F SPRR2F ELISA kit kit SpeciesHuman SpeciesHuman SpeciesHuman • Human Signal peptide CUB and EGF like domain • Human SLAIN motif containing protein 2 SLAIN2 ELISA kit • Human Small proline rich protein 2G SPRR2G ELISA kit containing protein 2 SCUBE2 ELISA kit SpeciesHuman SpeciesHuman SpeciesHuman • Human Signal peptide CUB and EGF like domain • Human SLAM family member 5 CD84 ELISA kit • Human Small proline rich protein 3 SPRR3 ELISA kit containing protein -

Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza a Virus (H7N9)
Article Proteomics Analysis of Cellular Proteins Co-Immunoprecipitated with Nucleoprotein of Influenza A Virus (H7N9) Ningning Sun 1,:, Wanchun Sun 2,:, Shuiming Li 3, Jingbo Yang 1, Longfei Yang 1, Guihua Quan 1, Xiang Gao 1, Zijian Wang 1, Xin Cheng 1, Zehui Li 1, Qisheng Peng 2,* and Ning Liu 1,* Received: 26 August 2015 ; Accepted: 22 October 2015 ; Published: 30 October 2015 Academic Editor: David Sheehan 1 Central Laboratory, Jilin University Second Hospital, Changchun 130041, China; [email protected] (N.S.); [email protected] (J.Y.); [email protected] (L.Y.); [email protected] (G.Q.); [email protected] (X.G.); [email protected] (Z.W.); [email protected] (X.C.); [email protected] (Z.L.) 2 Key Laboratory of Zoonosis Research, Ministry of Education, Institute of Zoonosis, Jilin University, Changchun 130062, China; [email protected] 3 College of Life Sciences, Shenzhen University, Shenzhen 518057, China; [email protected] * Correspondence: [email protected] (Q.P.); [email protected] (N.L.); Tel./Fax: +86-431-8879-6510 (Q.P. & N.L.) : These authors contributed equally to this work. Abstract: Avian influenza A viruses are serious veterinary pathogens that normally circulate among avian populations, causing substantial economic impacts. Some strains of avian influenza A viruses, such as H5N1, H9N2, and recently reported H7N9, have been occasionally found to adapt to humans from other species. In order to replicate efficiently in the new host, influenza viruses have to interact with a variety of host factors. In the present study, H7N9 nucleoprotein was transfected into human HEK293T cells, followed by immunoprecipitated and analyzed by proteomics approaches. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

PDF Datasheet
Product Datasheet FAM214A Antibody NBP1-83797 Unit Size: 0.1 ml Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews, Research Tools and Images at: www.novusbio.com/NBP1-83797 Updated 9/16/2020 v.20.1 Earn rewards for product reviews and publications. Submit a publication at www.novusbio.com/publications Submit a review at www.novusbio.com/reviews/destination/NBP1-83797 Page 1 of 2 v.20.1 Updated 9/16/2020 NBP1-83797 FAM214A Antibody Product Information Unit Size 0.1 ml Concentration Concentrations vary lot to lot. See vial label for concentration. If unlisted please contact technical services. Storage Store at 4C short term. Aliquot and store at -20C long term. Avoid freeze-thaw cycles. Clonality Polyclonal Preservative 0.02% Sodium Azide Isotype IgG Purity Immunogen affinity purified Buffer PBS (pH 7.2) and 40% Glycerol Product Description Host Rabbit Gene ID 56204 Gene Symbol FAM214A Species Human Specificity/Sensitivity Specificity of human FAM214A antibody verified on a Protein Array containing target protein plus 383 other non-specific proteins. Immunogen This antibody was developed against Recombinant Protein corresponding to amino acids: TNEGKIRLKPETPRSETCISNDFYSHMPVGETNPLIGSLLQERQDVIARIAQHLE HIDPTASHIPRQSFNMHDSSSVASKVFRSSYEDKNLLKKNKDESSVSISHT Product Application Details Applications Immunohistochemistry, Immunohistochemistry-Paraffin Recommended Dilutions Immunohistochemistry 1:20 - 1:50, Immunohistochemistry-Paraffin 1:20 - 1:50 Application Notes For IHC-Paraffin, HIER pH 6 retrieval is recommended. Immunocytochemistry/Immunofluorescence Fixation Permeabilization: Use PFA/Triton X-100. Images Immunohistochemistry-Paraffin: FAM214A Antibody [NBP1-83797] - Staining of human stomach shows strong cytoplasmic and membranous positivity in glandular cells. Novus Biologicals USA Bio-Techne Canada 10730 E. -

PRODUCT SPECIFICATION Prest Antigen FAM214A Product
PrEST Antigen FAM214A Product Datasheet PrEST Antigen PRODUCT SPECIFICATION Product Name PrEST Antigen FAM214A Product Number APrEST81419 Gene Description family with sequence similarity 214, member A Alternative Gene FLJ10980, KIAA1370 Names Corresponding Anti-FAM214A (HPA040180) Antibodies Description Recombinant protein fragment of Human FAM214A Amino Acid Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: EVLRTTLKHSNVWRKHNFHSLDGTSTRAFHPQTGLPLLSSPVPQRKTQSG CFDLDSSLLHLKSFSSRSPRPCLNIEDDPDIHEKPFLSSSAPPI Fusion Tag N-terminal His6ABP (ABP = Albumin Binding Protein derived from Streptococcal Protein G) Expression Host E. coli Purification IMAC purification Predicted MW 28 kDa including tags Usage Suitable as control in WB and preadsorption assays using indicated corresponding antibodies. Purity >80% by SDS-PAGE and Coomassie blue staining Buffer PBS and 1M Urea, pH 7.4. Unit Size 100 µl Concentration Lot dependent Storage Upon delivery store at -20°C. Avoid repeated freeze/thaw cycles. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

C6cc08797c1.Pdf
Electronic Supplementary Material (ESI) for Chemical Communications. This journal is © The Royal Society of Chemistry 2017 Supplementary Information for Competition-based, quantitative chemical proteomics in breast cancer cells identifies new target profiles for sulforaphane James A. Clulow,a,‡ Elisabeth M. Storck,a,‡ Thomas Lanyon-Hogg,a,‡ Karunakaran A. Kalesh,a Lyn Jonesb and Edward W. Tatea,* a Department of Chemistry, Imperial College London, London, UK, SW7 2AZ b Worldwide Medicinal Chemistry, Pfizer, 200 Cambridge Park Drive, MA 02140, USA ‡ Authors share equal contribution * corresponding author, email: [email protected] Table of Contents 1 Supporting Figures..........................................................................................................................3 2 Supporting Tables.........................................................................................................................17 2.1 Supporting Table S1. Incorporation validation for the R10K8 label in the 'spike-in' SILAC proteome of the MCF7 cell line........................................................................................................17 2.2 Supporting Table S2. Incorporation validation for the R10K8 label in the 'spike-in' SILAC proteome of the MDA-MB-231 cell line ...........................................................................................17 2.3 Supporting Table S3. High- and medium- confidence targets of sulforaphane in the MCF7 cell line..............................................................................................................................................17