Cnidaria: Anthozoa) Comprehensive Phylogenetic K
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Coelenterata: Anthozoa), with Diagnoses of New Taxa
PROC. BIOL. SOC. WASH. 94(3), 1981, pp. 902-947 KEY TO THE GENERA OF OCTOCORALLIA EXCLUSIVE OF PENNATULACEA (COELENTERATA: ANTHOZOA), WITH DIAGNOSES OF NEW TAXA Frederick M. Bayer Abstract.—A serial key to the genera of Octocorallia exclusive of the Pennatulacea is presented. New taxa introduced are Olindagorgia, new genus for Pseudopterogorgia marcgravii Bayer; Nicaule, new genus for N. crucifera, new species; and Lytreia, new genus for Thesea plana Deich- mann. Ideogorgia is proposed as a replacement ñame for Dendrogorgia Simpson, 1910, not Duchassaing, 1870, and Helicogorgia for Hicksonella Simpson, December 1910, not Nutting, May 1910. A revised classification is provided. Introduction The key presented here was an essential outgrowth of work on a general revisión of the octocoral fauna of the western part of the Atlantic Ocean. The far-reaching zoogeographical affinities of this fauna made it impossible in the course of this study to ignore genera from any part of the world, and it soon became clear that many of them require redefinition according to modern taxonomic standards. Therefore, the type-species of as many genera as possible have been examined, often on the basis of original type material, and a fully illustrated generic revisión is in course of preparation as an essential first stage in the redescription of western Atlantic species. The key prepared to accompany this generic review has now reached a stage that would benefit from a broader and more objective testing under practical conditions than is possible in one laboratory. For this reason, and in order to make the results of this long-term study available, even in provisional form, not only to specialists but also to the growing number of ecologists, biochemists, and physiologists interested in octocorals, the key is now pre- sented in condensed form with minimal illustration. -
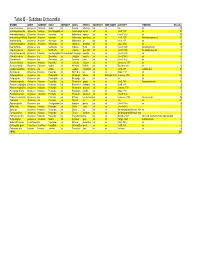
Table B – Subclass Octocorallia
Table B – Subclass Octocorallia BINOMEN ORDER SUBORDER FAMILY SUBFAMILY GENUS SPECIES SUBSPECIES COMN_NAMES AUTHORITY SYNONYMS #Records Acanella arbuscula Alcyonacea Calcaxonia Isididae n/a Acanella arbuscula n/a n/a n/a n/a 59 Acanthogorgia armata Alcyonacea Holaxonia Acanthogorgiidae n/a Acanthogorgia armata n/a n/a Verrill, 1878 n/a 95 Anthomastus agassizii Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus agassizii n/a n/a (Verrill, 1922) n/a 35 Anthomastus grandiflorus Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus grandiflorus n/a n/a Verrill, 1878 Anthomastus purpureus 37 Anthomastus sp. Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus sp. n/a n/a Verrill, 1878 n/a 1 Anthothela grandiflora Alcyonacea Scleraxonia Anthothelidae n/a Anthothela grandiflora n/a n/a (Sars, 1856) n/a 24 Capnella florida Alcyonacea n/a Nephtheidae n/a Capnella florida n/a n/a (Verrill, 1869) Eunephthya florida 44 Capnella glomerata Alcyonacea n/a Nephtheidae n/a Capnella glomerata n/a n/a (Verrill, 1869) Eunephthya glomerata 4 Chrysogorgia agassizii Alcyonacea Holaxonia Acanthogorgiidae Chrysogorgiidae Chrysogorgia agassizii n/a n/a (Verrill, 1883) n/a 2 Clavularia modesta Alcyonacea n/a Clavulariidae n/a Clavularia modesta n/a n/a (Verrill, 1987) n/a 6 Clavularia rudis Alcyonacea n/a Clavulariidae n/a Clavularia rudis n/a n/a (Verrill, 1922) n/a 1 Gersemia fruticosa Alcyonacea Alcyoniina Alcyoniidae n/a Gersemia fruticosa n/a n/a Marenzeller, 1877 n/a 3 Keratoisis flexibilis Alcyonacea Calcaxonia Isididae n/a Keratoisis flexibilis n/a n/a Pourtales, 1868 n/a 1 Lepidisis caryophyllia Alcyonacea n/a Isididae n/a Lepidisis caryophyllia n/a n/a Verrill, 1883 Lepidisis vitrea 13 Muriceides sp. -

Symbionts and Environmental Factors Related to Deep-Sea Coral Size and Health
Symbionts and environmental factors related to deep-sea coral size and health Erin Malsbury, University of Georgia Mentor: Linda Kuhnz Summer 2018 Keywords: deep-sea coral, epibionts, symbionts, ecology, Sur Ridge, white polyps ABSTRACT We analyzed video footage from a remotely operated vehicle to estimate the size, environmental variation, and epibiont community of three types of deep-sea corals (class Anthozoa) at Sur Ridge off the coast of central California. For all three of the corals, Keratoisis, Isidella tentaculum, and Paragorgia arborea, species type was correlated with the number of epibionts on the coral. Paragorgia arborea had the highest average number of symbionts, followed by Keratoisis. Epibionts were identified to the lowest possible taxonomic level and categorized as predators or commensalists. Around twice as many Keratoisis were found with predators as Isidella tentaculum, while no predators were found on Paragorgia arborea. Corals were also measured from photos and divided into size classes for each type based on natural breaks. The northern sites of the mound supported larger Keratoisis and Isidella tentaculum than the southern portion, but there was no relationship between size and location for Paragorgia arborea. The northern sites of Sur Ridge were also the only place white polyps were found. These polyps were seen mostly on Keratoisis, but were occasionally found on the skeletons of Isidella tentaculum and even Lillipathes, an entirely separate subclass of corals from Keratoisis. Overall, although coral size appears to be impacted by 1 environmental variables and location for Keratoisis and Isidella tentaculum, the presence of symbionts did not appear to correlate with coral size for any of the coral types. -

Deep-Sea Origin and In-Situ Diversification of Chrysogorgiid Octocorals
Deep-Sea Origin and In-Situ Diversification of Chrysogorgiid Octocorals Eric Pante1*¤, Scott C. France1, Arnaud Couloux2, Corinne Cruaud2, Catherine S. McFadden3, Sarah Samadi4, Les Watling5,6 1 Department of Biology, University of Louisiana at Lafayette, Lafayette, Louisiana, United States of America, 2 GENOSCOPE, Centre National de Se´quenc¸age, Evry, France, 3 Department of Biology, Harvey Mudd College, Claremont, California, United States of America, 4 De´partement Syste´matique et Evolution, UMR 7138 UPMC-IRD-MNHN- CNRS (UR IRD 148), Muse´um national d’Histoire naturelle, Paris, France, 5 Department of Biology, University of Hawaii at Manoa, Honolulu, Hawaii, United States of America, 6 Darling Marine Center, University of Maine, Walpole, Maine, United States of America Abstract The diversity, ubiquity and prevalence in deep waters of the octocoral family Chrysogorgiidae Verrill, 1883 make it noteworthy as a model system to study radiation and diversification in the deep sea. Here we provide the first comprehensive phylogenetic analysis of the Chrysogorgiidae, and compare phylogeny and depth distribution. Phylogenetic relationships among 10 of 14 currently-described Chrysogorgiidae genera were inferred based on mitochondrial (mtMutS, cox1) and nuclear (18S) markers. Bathymetric distribution was estimated from multiple sources, including museum records, a literature review, and our own sampling records (985 stations, 2345 specimens). Genetic analyses suggest that the Chrysogorgiidae as currently described is a polyphyletic family. Shallow-water genera, and two of eight deep-water genera, appear more closely related to other octocoral families than to the remainder of the monophyletic, deep-water chrysogorgiid genera. Monophyletic chrysogorgiids are composed of strictly (Iridogorgia Verrill, 1883, Metallogorgia Versluys, 1902, Radicipes Stearns, 1883, Pseudochrysogorgia Pante & France, 2010) and predominantly (Chrysogorgia Duchassaing & Michelotti, 1864) deep-sea genera that diversified in situ. -

Author's Accepted Manuscript
Author’s Accepted Manuscript This is the accepted version of the following article: Rakka, M., Bilan, M., Godinho, A., Movilla, J., Orejas, C., & Carreiro-Silva, M. (2019). First description of polyp bailout in cold-water octocorals under aquaria maintenance. Coral Reefs, 38(1), 15-20, which has been published in final form at https://doi.org/10.1007/s00338-018-01760- x. This article may be used for non-commercial purposes in accordance with Springer Terms and Conditions for Use of Self-Archived Versions 1 First description of polyp bail-out in cold-water octocorals under aquaria maintenance Maria Rakka1,2,3, Meri Bilan1,2,3, Antonio Godinho1,2,3,Juancho Movilla4,5, Covadonga Orejas4, Marina Carreiro-Silva1,2,3 1 MARE – Marine and Environmental Sciences Centre and Centre of the Institute of Marine Research 2 IMAR – University of the Azores, Rua Frederico Machado 4, 9901-862 Horta, Portugal 3 OKEANOS Research Unit, Faculty of Science and Technology, University of the Azores, 9901-862, Horta, Portugal 4 Instituto Español de Oceanografía, Centro Oceanográfico de Baleares, Moll de Ponent s/n, 07015 Palma, Spain 5 Instituto de Ciencias del Mar (ICM-CSIC). Passeig Maritim de la Barceloneta 37-49, 08003 Barcelona, Spain Corresponding author: Maria Rakka, [email protected], +351915407062 2 Abstract Cnidarians, characterized by high levels of plasticity, exhibit remarkable mechanisms to withstand or escape unfavourable conditions including reverse development which describes processes of transformation of adult stages into early developmental stages with higher mobility. Polyp bail-out is a stress-escape response common among scleractinian species, consisting of massive detachment of live polyps and subsequent death of the mother colony. -

Comprehensive Phylogenomic Analyses Resolve Cnidarian Relationships and the Origins of Key Organismal Traits
Comprehensive phylogenomic analyses resolve cnidarian relationships and the origins of key organismal traits Ehsan Kayal1,2, Bastian Bentlage1,3, M. Sabrina Pankey5, Aki H. Ohdera4, Monica Medina4, David C. Plachetzki5*, Allen G. Collins1,6, Joseph F. Ryan7,8* Authors Institutions: 1. Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution 2. UPMC, CNRS, FR2424, ABiMS, Station Biologique, 29680 Roscoff, France 3. Marine Laboratory, university of Guam, UOG Station, Mangilao, GU 96923, USA 4. Department of Biology, Pennsylvania State University, University Park, PA, USA 5. Department of Molecular, Cellular and Biomedical Sciences, University of New Hampshire, Durham, NH, USA 6. National Systematics Laboratory, NOAA Fisheries, National Museum of Natural History, Smithsonian Institution 7. Whitney Laboratory for Marine Bioscience, University of Florida, St Augustine, FL, USA 8. Department of Biology, University of Florida, Gainesville, FL, USA PeerJ Preprints | https://doi.org/10.7287/peerj.preprints.3172v1 | CC BY 4.0 Open Access | rec: 21 Aug 2017, publ: 21 Aug 20171 Abstract Background: The phylogeny of Cnidaria has been a source of debate for decades, during which nearly all-possible relationships among the major lineages have been proposed. The ecological success of Cnidaria is predicated on several fascinating organismal innovations including symbiosis, colonial body plans and elaborate life histories, however, understanding the origins and subsequent diversification of these traits remains difficult due to persistent uncertainty surrounding the evolutionary relationships within Cnidaria. While recent phylogenomic studies have advanced our knowledge of the cnidarian tree of life, no analysis to date has included genome scale data for each major cnidarian lineage. Results: Here we describe a well-supported hypothesis for cnidarian phylogeny based on phylogenomic analyses of new and existing genome scale data that includes representatives of all cnidarian classes. -

Deep-Sea Life Issue 8, November 2016 Cruise News Going Deep: Deepwater Exploration of the Marianas by the Okeanos Explorer
Deep-Sea Life Issue 8, November 2016 Welcome to the eighth edition of Deep-Sea Life: an informal publication about current affairs in the world of deep-sea biology. Once again we have a wealth of contributions from our fellow colleagues to enjoy concerning their current projects, news, meetings, cruises, new publications and so on. The cruise news section is particularly well-endowed this issue which is wonderful to see, with voyages of exploration from four of our five oceans from the Arctic, spanning north east, west, mid and south Atlantic, the north-west Pacific, and the Indian Ocean. Just imagine when all those data are in OBIS via the new deep-sea node…! (see page 24 for more information on this). The photo of the issue makes me smile. Angelika Brandt from the University of Hamburg, has been at sea once more with her happy-looking team! And no wonder they look so pleased with themselves; they have collected a wonderful array of life from one of the very deepest areas of our ocean in order to figure out more about the distribution of these abyssal organisms, and the factors that may limit their distribution within this region. Read more about the mission and their goals on page 5. I always appreciate feedback regarding any aspect of the publication, so that it may be improved as we go forward. Please circulate to your colleagues and students who may have an interest in life in the deep, and have them contact me if they wish to be placed on the mailing list for this publication. -

Guide to the Identification of Precious and Semi-Precious Corals in Commercial Trade
'l'llA FFIC YvALE ,.._,..---...- guide to the identification of precious and semi-precious corals in commercial trade Ernest W.T. Cooper, Susan J. Torntore, Angela S.M. Leung, Tanya Shadbolt and Carolyn Dawe September 2011 © 2011 World Wildlife Fund and TRAFFIC. All rights reserved. ISBN 978-0-9693730-3-2 Reproduction and distribution for resale by any means photographic or mechanical, including photocopying, recording, taping or information storage and retrieval systems of any parts of this book, illustrations or texts is prohibited without prior written consent from World Wildlife Fund (WWF). Reproduction for CITES enforcement or educational and other non-commercial purposes by CITES Authorities and the CITES Secretariat is authorized without prior written permission, provided the source is fully acknowledged. Any reproduction, in full or in part, of this publication must credit WWF and TRAFFIC North America. The views of the authors expressed in this publication do not necessarily reflect those of the TRAFFIC network, WWF, or the International Union for Conservation of Nature (IUCN). The designation of geographical entities in this publication and the presentation of the material do not imply the expression of any opinion whatsoever on the part of WWF, TRAFFIC, or IUCN concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The TRAFFIC symbol copyright and Registered Trademark ownership are held by WWF. TRAFFIC is a joint program of WWF and IUCN. Suggested citation: Cooper, E.W.T., Torntore, S.J., Leung, A.S.M, Shadbolt, T. and Dawe, C. -

Taxonomy and Phylogenetic Relationships of the Coral Genera Australomussa and Parascolymia (Scleractinia, Lobophylliidae)
Contributions to Zoology, 83 (3) 195-215 (2014) Taxonomy and phylogenetic relationships of the coral genera Australomussa and Parascolymia (Scleractinia, Lobophylliidae) Roberto Arrigoni1, 7, Zoe T. Richards2, Chaolun Allen Chen3, 4, Andrew H. Baird5, Francesca Benzoni1, 6 1 Dept. of Biotechnology and Biosciences, University of Milano-Bicocca, 20126, Milan, Italy 2 Aquatic Zoology, Western Australian Museum, 49 Kew Street, Welshpool, WA 6106, Australia 3Biodiversity Research Centre, Academia Sinica, Nangang, Taipei 115, Taiwan 4 Institute of Oceanography, National Taiwan University, Taipei 106, Taiwan 5 ARC Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD 4811, Australia 6 Institut de Recherche pour le Développement, UMR227 Coreus2, 101 Promenade Roger Laroque, BP A5, 98848 Noumea Cedex, New Caledonia 7 E-mail: [email protected] Key words: COI, evolution, histone H3, Lobophyllia, Pacific Ocean, rDNA, Symphyllia, systematics, taxonomic revision Abstract Molecular phylogeny of P. rowleyensis and P. vitiensis . 209 Utility of the examined molecular markers ....................... 209 Novel micromorphological characters in combination with mo- Acknowledgements ...................................................................... 210 lecular studies have led to an extensive revision of the taxonomy References ...................................................................................... 210 and systematics of scleractinian corals. In the present work, we Appendix ....................................................................................... -

Warm Seawater Temperature Promotes Substrate Colonization by the Blue Coral, Heliopora Coerulea
Warm seawater temperature promotes substrate colonization by the blue coral, Heliopora coerulea Author Christine Guzman, Michael Atrigenio, Chuya Shinzato, Porfirio Alino, Cecilia Conaco journal or PeerJ publication title volume 7 page range e7785 year 2019-09-27 Publisher PeerJ Rights (C) 2019 Guzman et al. Author's flag publisher URL http://id.nii.ac.jp/1394/00001286/ doi: info:doi/10.7717/peerj.7785 Creative Commons Attribution 4.0 International(https://creativecommons.org/licenses/by/4.0/) Warm seawater temperature promotes substrate colonization by the blue coral, Heliopora coerulea Christine Guzman1,2, Michael Atrigenio1, Chuya Shinzato3, Porfirio Aliño1 and Cecilia Conaco1 1 Marine Science Institute, College of Science, University of the Philippines Diliman, Quezon City, Philippines 2 Evolutionary Neurobiology Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Okinawa, Japan 3 Department of Marine Bioscience, Atmosphere and Ocean Research Institute, The University of Tokyo, Kashiwa-shi, Chiba, Japan ABSTRACT Background: Heliopora coerulea, the blue coral, is a reef building octocoral that is reported to have a higher optimum temperature for growth compared to most scleractinian corals. This octocoral has been observed to grow over both live and dead scleractinians and to dominate certain reefs in the Indo-Pacific region. The molecular mechanisms underlying the ability of H. coerulea to tolerate warmer seawater temperatures and to effectively compete for space on the substrate remain to be elucidated. Methods: In this study, we subjected H. coerulea colonies to various temperatures for up to 3 weeks. The growth and photosynthetic efficiency rates of the coral colonies were measured. We then conducted pairwise comparisons of gene expression among the different coral tissue regions to identify genes and pathways that are expressed under different temperature conditions. -

Count Or Pointcount: Is Percent Octocoral Cover an Adequate Proxy for Octocoral Abundance? Matthew .J Lybolt University of South Florida
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School 4-4-2003 Count or Pointcount: Is Percent Octocoral Cover an Adequate Proxy for Octocoral Abundance? Matthew .J Lybolt University of South Florida Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the American Studies Commons Scholar Commons Citation Lybolt, Matthew J., "Count or Pointcount: Is Percent Octocoral Cover an Adequate Proxy for Octocoral Abundance?" (2003). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/1422 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. COUNT OR POINTCOUNT: IS PERCENT OCTOCORAL COVER AN ADEQUATE PROXY FOR OCTOCORAL ABUNDANCE? by MATTHEW J. LYBOLT A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science Department of Biological Oceanography College of Marine Science University of South Florida Major Professor: Pamela Hallock Muller, Ph.D. Walter C. Jaap, B.S. James W. Porter, Ph.D. George Yanev, Ph.D. Date of Approval: 4 April 2003 Keywords: Octocoral, Gorgonia ventalina, Florida Keys, Percent Cover, Hurricane Georges © Copyright 2003, Matthew Lybolt Acknowledgements I would like to thank and acknowledge my major professor Dr. Pamela Hallock Muller for valuable input and much appreciated latitude. With her guidance, my skill as a writer and my capacity as a scientist have improved greatly. I would like to thank my committee members Walter Jaap, Dr. -

The Genetic Identity of Dinoflagellate Symbionts in Caribbean Octocorals
Coral Reefs (2004) 23: 465-472 DOI 10.1007/S00338-004-0408-8 REPORT Tamar L. Goulet • Mary Alice CofFroth The genetic identity of dinoflagellate symbionts in Caribbean octocorals Received: 2 September 2002 / Accepted: 20 December 2003 / Published online: 29 July 2004 © Springer-Verlag 2004 Abstract Many cnidarians (e.g., corals, octocorals, sea Introduction anemones) maintain a symbiosis with dinoflagellates (zooxanthellae). Zooxanthellae are grouped into The cornerstone of the coral reef ecosystem is the sym- clades, with studies focusing on scleractinian corals. biosis between cnidarians (e.g., corals, octocorals, sea We characterized zooxanthellae in 35 species of Caribbean octocorals. Most Caribbean octocoral spe- anemones) and unicellular dinoñagellates commonly called zooxanthellae. Studies of zooxanthella symbioses cies (88.6%) hosted clade B zooxanthellae, 8.6% have previously been hampered by the difficulty of hosted clade C, and one species (2.9%) hosted clades B and C. Erythropodium caribaeorum harbored clade identifying the algae. Past techniques relied on culturing and/or identifying zooxanthellae based on their free- C and a unique RFLP pattern, which, when se- swimming form (Trench 1997), antigenic features quenced, fell within clade C. Five octocoral species (Kinzie and Chee 1982), and cell architecture (Blank displayed no zooxanthella cladal variation with depth. 1987), among others. These techniques were time-con- Nine of the ten octocoral species sampled throughout suming, required a great deal of expertise, and resulted the Caribbean exhibited no regional zooxanthella cla- in the differentiation of only a small number of zoo- dal differences. The exception, Briareum asbestinum, xanthella species. Molecular techniques amplifying had some colonies from the Dry Tortugas exhibiting zooxanthella DNA encoding for the small and large the E.