Development 129
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

The Eyes Have It
news and views flow of heat and matter in the interior2. This Irifune and Isshiki1 have shown such transition to higher pressures (as can be seen flow, in turn, drives the tectonic evolution artificial separation to be misleading by in Irifune and Isshiki’s Fig. 3 on page 704), so of the surface, including the occurrence of experimentally demonstrating Mg–Fe ex- the onset of the transition in pyrolite is post- volcanism and seismicity. change between a, b, garnet and clino- poned to greater depths relative to that in Olivine, a solid solution of forsterite pyroxene. We have known5 for some time Fo89. On the other side of the phase change, b (Mg2SiO4) and fayalite (Fe2SiO4), has the that the proportions of these minerals vary does not partition Fe into garnet as strong- a b approximate composition (Mg0.89 Fe0.11)2SiO4 with depth, as shown by the bold white lines ly as does , so is not initially Mg-enriched, (called forsterite-89 or Fo89) in samples from in Fig. 1, but Irifune and Isshiki have now and the completion of the a–b transition the shallow mantle. It transforms from the measured the accompanying variations in is not postponed relative to Fo89. The net olivine (a) phase to wadsleyite (b), changing the compositions of coexisting minerals, effect of postponing onset but not co8m- again to ringwoodite (g) at greater depths shown by the colours. In isolated Fo89 pletion is to concentrate the transition into and eventually breaking down to a mixture of olivine, mineral compositions must remain a narrower range of depths, decreasing the silicate perovskite and magnesiowüstite. -

The Upper Mantle and Transition Zone
The Upper Mantle and Transition Zone Daniel J. Frost* DOI: 10.2113/GSELEMENTS.4.3.171 he upper mantle is the source of almost all magmas. It contains major body wave velocity structure, such as PREM (preliminary reference transitions in rheological and thermal behaviour that control the character Earth model) (e.g. Dziewonski and Tof plate tectonics and the style of mantle dynamics. Essential parameters Anderson 1981). in any model to describe these phenomena are the mantle’s compositional The transition zone, between 410 and thermal structure. Most samples of the mantle come from the lithosphere. and 660 km, is an excellent region Although the composition of the underlying asthenospheric mantle can be to perform such a comparison estimated, this is made difficult by the fact that this part of the mantle partially because it is free of the complex thermal and chemical structure melts and differentiates before samples ever reach the surface. The composition imparted on the shallow mantle by and conditions in the mantle at depths significantly below the lithosphere must the lithosphere and melting be interpreted from geophysical observations combined with experimental processes. It contains a number of seismic discontinuities—sharp jumps data on mineral and rock properties. Fortunately, the transition zone, which in seismic velocity, that are gener- extends from approximately 410 to 660 km, has a number of characteristic ally accepted to arise from mineral globally observed seismic properties that should ultimately place essential phase transformations (Agee 1998). These discontinuities have certain constraints on the compositional and thermal state of the mantle. features that correlate directly with characteristics of the mineral trans- KEYWORDS: seismic discontinuity, phase transformation, pyrolite, wadsleyite, ringwoodite formations, such as the proportions of the transforming minerals and the temperature at the discontinu- INTRODUCTION ity. -

Phase Transitions in Earth's Mantle and Mantle Mineralogy
Phase transitions in Earth’s Mantle and Mantle Mineralogy Upper Mantle Minerals: Olivine, Orthopyroxene, Clinopyroxene and Garnet ~13.5 GPa: Olivine Æ Wadsrlyite (DE) transition (ONSET TRANSITION ZONE) ~15.5 GPa: Pyroxene component gradually dissolve into garnet structure, resulting in the completion of pyroxene-majorite transformation >20 GPa: High CaO content in majorite is unfavorable at high pressure, leading to the formation of CaSiO3 perovskite ~24 GPa: Division of transition zone and lower mantle. Sharp transition silicate spinel to ferromagnesium silicate perovskite and magnesiowustite >24 GPa: Most of Al2O3 resides in majorite at transition zone pressures, a transformation from Majorite to Al-bearing orthorhombic perovskite completes at pressure higher than that of post-spinel transformation Lower Mantle Minerals: Orthorhobic perovskite, Magnesiowustite, CaSiO3 perovskite ~27 GPa: Transformation of Al and Si rich basalt to perovskite lithology with assemblage of Al-bearing perovskite, CaSiO3, stishovite and Al-phases Upper Mantle: olivine, garnet and pyroxene Transition zone: olivine (a-phase) transforms to wadsleyite (b-phase) then to spinel structure (g-phase) and finally to perovskite + magnesio-wüstite. Transformations occur at P and T conditions similar to 410, 520 and 660 km seismic discontinuities Xenoliths: (mantle fragments brought to surface in lavas) 60% Olivine + 40 % Pyroxene + some garnet Images removed due to copyright considerations. Garnet: A3B2(SiO4)3 Majorite FeSiO4, (Mg,Fe)2 SiO4 Germanates (Co-, Ni- and Fe- -

Time-Varying Subduction and Rollback Velocities in Slab Stagnation
Phase transformation of harzburgite and stagnant slab in the mantle transition zone Y. Zh ang 1,2, Y. Wu1, Y. Wang2, Z. Jin1, C. R. Bina3 1State Key Laboratory of Geological Processes and Mineral Resources (GPMR), China University of Geosciences, Wuhan, China 2Center for Advanced Radiation Sources, The University of Chicago, Chicago, IL, USA 3Dept. of Earth and Planetary Sciences, Northwestern University, Evanston, IL, USA Subducted materials play an important role in affecting chemical composition and structure of the mantle transition zone. Harzburgite is generally accepted as an important part of subducting slabs, overlain by a layer of basalt. Seismic tomography studies have detected widespread fast anomalies in the mantle transition zone (MTZ) and lower mantle around the circum-Pacific and the Mediterranean region. These anomalies have been interpreted as stagnant oceanic lithosphere materials as these regions are closely associated with subduction zones, where the oceanic lithosphere plunges deeply into the MTZ. However, experimental studies on the phase transformations of harzburgite have been very limited, and no experimental studies have been conducted on the physical properties of harzburgite under MTZ conditions. In this study, we conducted high-temperature and high-pressure experiments, using a 1000-ton Kawai-type multi-anvil apparatus at GPMR, on a natural harzburgite at 14.1-24.2 GPa and 1473-1673 K. At 1673 K, harzburgite transformed to wadsleyite + garnet + clinopyroxene below 19 GPa and further decomposed into an assemblage of ringwoodite + garnet + stishovite above 20 GPa. Some amounts of akimotoite were produced at still higher pressures (22-23 GPa), and finally perovskite and magnesiowustite were found to coexist with garnet at 24.2 GPa. -

Geochemistry
GEOCHEMISTRY “Structure & Composition of the Earth” (M.Sc. Sem IV) Shekhar Assistant Professor Department of Geology Patna Science College Patna University E-mail: [email protected] Mob: +91-7004784271 Structure of the Earth • Earth structure and it’s composition is the essential component of Geochemistry. • Seismology is the main tool for the determination of the Earth’s interior. • Interpretation of the property is based on the behaviour of two body waves travelling within the interior. Structure of the Earth Variation in seismic body-wave paths, which in turn represents the variation in properties of the earth’s interior. Structure of the Earth Based on seismic data, Earth is broadly divided into- i. The Crust ii. The Mantle iii. The Core *Figure courtesy Geologycafe.com Density, Pressure & Temperature variation with Depth *From Bullen, An introduction to the theory of seismology. Courtesy of Cambridge Cambridge University Press *Figure courtesy Brian Mason: Principle of Geochemistry The crust • The crust is the outermost layer of the earth. • It consist 0.5-1.0 per cent of the earth’s volume and less than 1 per cent of Earth’s mass. • The average density is about “2.7 g/cm3” (average density of the earth is 5.51 g/cm³). • The crust is differentiated into- i) Oceanic crust ii) Continental crust The Oceanic crust • Covers approx. ~ 70% of the Earth’s surface area. • Average thickness ~ 6km (~4km at MOR : ~10km at volcanic plateau) • Mostly mafic in nature. • Relatively younger in age. The Oceanic crust Seismic study shows layered structural arrangement. - Seawater - Sediments - Basaltic layer - Gabbroic layer *Figure courtesy W.M.White: Geochemistry The Continental crust • Heterogeneous in nature. -

Crystal Structures of Minerals in the Lower Mantle
6 Crystal Structures of Minerals in the Lower Mantle June K. Wicks and Thomas S. Duffy ABSTRACT The crystal structures of lower mantle minerals are vital components for interpreting geophysical observations of Earth’s deep interior and in understanding the history and composition of this complex and remote region. The expected minerals in the lower mantle have been inferred from high pressure‐temperature experiments on mantle‐relevant compositions augmented by theoretical studies and observations of inclusions in natural diamonds of deep origin. While bridgmanite, ferropericlase, and CaSiO3 perovskite are expected to make up the bulk of the mineralogy in most of the lower mantle, other phases such as SiO2 polymorphs or hydrous silicates and oxides may play an important subsidiary role or may be regionally important. Here we describe the crystal structure of the key minerals expected to be found in the deep mantle and discuss some examples of the relationship between structure and chemical and physical properties of these phases. 6.1. IntrODUCTION properties of lower mantle minerals [Duffy, 2005; Mao and Mao, 2007; Shen and Wang, 2014; Ito, 2015]. Earth’s lower mantle, which spans from 660 km depth The crystal structure is the most fundamental property to the core‐mantle boundary (CMB), encompasses nearly of a mineral and is intimately related to its major physical three quarters of the mass of the bulk silicate Earth (crust and chemical characteristics, including compressibility, and mantle). Our understanding of the mineralogy and density, -

2.02 Seismological Constraints Upon Mantle Composition C
2.02 Seismological Constraints upon Mantle Composition C. R. Bina Northwestern University, Evanston, IL, USA 2.02.1 INTRODUCTION 39 2.02.1.1 General Considerations 39 2.02.1.2 Bulk Sound Velocity 40 2.02.1.3 Acoustic Methods 40 2.02.2 UPPER-MANTLE BULK COMPOSITION 41 2.02.2.1 Overview 41 2.02.2.2 Velocity Contrasts 42 2.02.2.3 Discontinuity Topography 42 2.02.2.4 Sharpness 43 2.02.2.5 Broadening and Bifurcation 43 2.02.3 UPPER-MANTLE HETEROGENEITY 45 2.02.3.1 Subducted Basalts 45 2.02.3.2 Plume Origins 46 2.02.4 LOWER-MANTLE BULK COMPOSITION 48 2.02.4.1 Bulk Fitting 48 2.02.4.2 Depthwise Fitting 49 2.02.5 LOWER-MANTLE HETEROGENEITY 51 2.02.5.1 Overview 51 2.02.5.2 Subducted Oceanic Crust 52 2.02.6 SUMMARY 54 REFERENCES 56 2.02.1 INTRODUCTION and lower mantle? What light can they shed upon 2.02.1.1 General Considerations the nature of velocity heterogeneities in both the upper and lower mantle? It is these questions that Direct sampling of mantle rocks and minerals is we shall seek to address in this chapter. limited to tectonic slices emplaced at the surface Most seismological constraints on mantle com- (see Chapter 2.04), smaller xenoliths transported position are derived by comparison of values of upwards by magmatic processes (see Chapter seismic wave velocities inferred for particular 2.05), and still smaller inclusions in such far- regions within the Earth to the values measured in traveled natural sample chambers as diamonds the laboratory for particular minerals or mineral (see Chapter 2.05). -

Confirming a Pyrolitic Lower Mantle Using Self-Consistent Pressure
PUBLICATIONS Journal of Geophysical Research: Solid Earth RESEARCH ARTICLE Confirming a pyrolitic lower mantle using self-consistent 10.1002/2016JB013062 pressure scales and new constraints Key Points: on CaSiO3 perovskite • Modeling the ρ and VΦ profiles of the lower mantle using self-consistent Ningyu Sun1, Zhu Mao1,2, Shuai Yan1, Xiang Wu3, Vitali B. Prakapenka4, and Jung-Fu Lin5,6 pressure scales and new constraints on Ca-Pv 1Laboratory of Seismology and Physics of Earth’s Interior, School of Earth and Planetary Sciences, University of Science and • Comparing with PREM, a pyrolitic 2 3 mantle is preferred over a chondritic Technology of China, Hefei, China, National Geophysics Observatory, Mengcheng, China, State Key Laboratory of 4 mantle Geological Processes and Mineral Resources, China University of Geosciences, Wuhan, China, Center for Advanced • The amount of Ca-Pv cannot be Radiation Sources, University of Chicago, Chicago, Illinois, USA, 5Department of Geological Sciences, Jackson School of constrained based on the density and Geosciences, University of Texas at Austin, Austin, Texas, USA, 6Center for High Pressure Science and Technology Advanced bulk sound velocity profiles as its Research, Shanghai, China elasticity lies close to PREM Abstract In this study, we have examined the lower mantle composition and mineralogy by modeling the Correspondence to: density (ρ), bulk sound velocity (VΦ), and dlnρ/dlnVΦ profiles of candidate lower mantle minerals using Z. Mao, literature and new experimental equation of state (EoS) results. For CaSiO3 perovskite, complimentary [email protected] synchrotron X-ray diffraction measurements in a laser-heated diamond anvil cell were conducted up to 156 GPa between 1200 K and 2600 K to provide more reliable constraints on the thermal EoS parameters. -

The Evolution and Distribution of Recycled Oceanic Crust in The
1 The evolution and distribution of recycled oceanic crust in the 2 Earth’s mantle: Insight from geodynamic models a b, a, c a 3 Jun Yan , Maxim D. Ballmer , and Paul J. Tackley a 4 Institute of Geophysics, ETH Zurich, Zurich, Switzerland b 5 Department of Earth Sciences, University College London, London, United Kingdom c 6 Earth-Life Science Institute, Tokyo Institute of Technology, Tokyo, Japan 7 Corresponding author: Jun Yan ([email protected]) 8 Highlights: 9 • Compositional mantle layering is robustly predicted despite whole-mantle convection. 10 • Basalt-enhanced reservoir forms in the MTZ, independent of mantle viscosity profile. 11 • Basalt fraction in the MTZ is laterally variable, ranging from ~30% to 50%. 12 • A layer beneath the MTZ displays significant (40%~80%) enrichment in harzburgite. 13 • The bulk-silicate Earth may be enriched in basalt relative to upper-mantle pyrolite. 14 Keywords: recycled oceanic crust, compositional mantle layering, chemical heterogeneity, 15 mantle transition zone, thermochemical convection 16 Abstract 17 A better understanding of the Earth’s compositional structure is needed to place the geochemical 18 record of surface rocks into the context of Earth accretion and evolution. Cosmochemical 19 constraints imply that lower-mantle rocks may be enriched in silica relative to upper-mantle 20 pyrolite, whereas geophysical observations support whole-mantle convection and mixing. To 21 resolve this discrepancy, it has been suggested that subducted mid-ocean ridge basalt (MORB) 22 segregates from subducted harzburgite to accumulate in the mantle transition zone (MTZ) and/or 23 the lower mantle. However, the key parameters that control basalt segregation and accumulation 24 remain poorly constrained. -

Phase Relations in MAFSH System up to 21 Gpa: Implications for Water Cycles in Martian Interior
minerals Article Phase Relations in MAFSH System up to 21 GPa: Implications for Water Cycles in Martian Interior Chaowen Xu 1,* and Toru Inoue 1,2,3 1 Geodynamics Research Center, Ehime University, 2-5 Bunkyo-cho, Matsuyama 790-8577, Japan; [email protected] 2 Department of Earth and Planetary Systems Science, Hiroshima University, 1-3-1 Kagamiyama, Higashi-Hiroshima 739-8526, Japan 3 Hiroshima Institute of Plate Convergence Region Research (HiPeR), Hiroshima University, Higashi-Hiroshima, Hiroshima 739-8526, Japan * Correspondence: [email protected]; Tel.: +81-050-3699-0952 Received: 3 August 2019; Accepted: 14 September 2019; Published: 16 September 2019 Abstract: To elucidate the water cycles in iron-rich Mars, we investigated the phase relation of a water-undersaturated (2 wt.%) analog of Martian mantle in simplified MgO-Al2O3-FeO-SiO2-H2O (MAFSH) system between 15 and 21 GPa at 900–1500 ◦C using a multi-anvil apparatus. Results showed that phase E coexisting with wadsleyite or ringwoodite was at least stable at 15–16.5 GPa and below 1050 ◦C. Phase D coexisted with ringwoodite at pressures higher than 16.5 GPa and temperatures below 1100 ◦C. The transition pressure of the loop at the wadsleyite-ringwoodite boundary shifted towards lower pressure in an iron-rich system compared with a hydrous pyrolite model of the Earth. Some evidence indicates that water once existed on the Martian surface on ancient Mars. The water present in the hydrous crust might have been brought into the deep interior by the convecting mantle. Therefore, water might have been transported to the deep Martian interior by hydrous minerals, such as phase E and phase D, in cold subduction plates. -
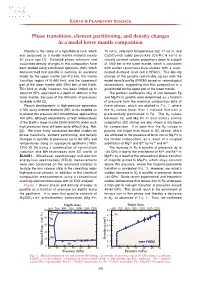
Phase Transitions, Element Partitioning, and Density Changes in a Model Lower Mantle Composition T. Irifune
Earth & Planetary Science Phase transitions, element partitioning, and density changes in a model lower mantle composition Pyrolite is the name of a hypothetical rock, which 75 vol%), (Mg,Fe)O ferropericlase (Fp; 17 vol%), and was proposed as a model mantle material nearly CaSiO 3-rich cubic perovskite (Ca-Pv; 8 vol%) in 50 years ago [1]. Detailed phase relations and virtually constant volume proportions down to a depth associated density changes in this composition have of 1200 km in the lower mantle, which is consistent been studied using a multianvil apparatus (MA), which with earlier reconnaissance studies with a laser- demonstrated that pyrolite is certainly an excellent heated diamond anvil cell (LHDAC). The density model for the upper mantle (3 0- 410 km), the mantle change of the pyrolite remarkably agrees with the transition region (41 0- 660 km), and the uppermost model density profile (PREM) based on seismological part of the lower mantle (66 0- 2900 km) of the Earth. observations, suggesting that this composition is a This kind of study, however, has been limited up to good model for the upper part of the lower mantle. about 30 GPa, equivalent to a depth of ~800 km in the The partition coefficients ( KD) of iron between Fp lower mantle, because of the limitation of pressures and Mg-Pv in pyrolite were determined as a function available in MA [2]. of pressure from the chemical composition data of Recent developments in high-pressure generation these phases, which are plotted in Fig. 2 , where in MA using sintered diamond (SD) anvils enabled us the KD values lower than 1 indicate that iron is to extend this pressure limit toward those approaching preferentially partitioned in Fp. -

1 Revision 3: Phase Transformation of Hydrous Ringwoodite to the Lower
Revision 3: Phase Transformation of Hydrous Ringwoodite to the Lower-Mantle Phases and the Formation of Dense Hydrous Silica Huawei Chen1, Kurt Leinenweber2, Vitali Prakapenka3, Martin Kunz4, Hans A. Bechtel4, Zhenxian Liu5 and Sang-Heon Shim1 1 School of Earth and Space Exploration, Arizona State University, Tempe, Arizona. 2 Eyring Materials Center, Arizona State University, Tempe, Arizona. 3 GeoSoilEnviroCars, University of Chicago, Chicago, Illinois. 4 Advanced Light Source Division, Lawrence Berkeley National Laboratory, Berkeley, California. 5 Geophysical Laboratory, Carnegie Institution of Washington, Washington, DC. Keywords: stishovite, ringwoodite, bridgmanite, periclase, water, mantle ABstract In order to understand the effects of H2O on the mineral phases forming under the pressure-temperature conditions of the lower mantle, we have conducted laser-heated diamond-anvil cell experiments on hydrous ringwoodite (Mg2SiO4 with 1.1wt% H2O) at pressures between 29 and 59 GPa and temperatures between 1200 and 2400 K. Our experimental results show that hydrous ringwoodite (hRw) converts to crystalline dense hydrous silica, stishovite (Stv) or CaCl2-type SiO2 (mStv), containing 1 wt% H2O together with Brd and MgO at the pressure-temperature conditions expected for shallow lower-mantle depths between approximately 660 to 1600 km. Considering lack of sign for melting in our experiments, our preferred interpretation of the observation is that Brd partially breaks down to dense hydrous silica and periclase (Pc), forming Brd + Pc + Stv mineralogy. Our experiments may provide an explanation for the enigmatic coexistence of Stv and Fp inclusions in lower-mantle diamonds. Introduction Lines of evidence support that the lower mantle has a similar chemical composition to the upper mantle (Kurnosov et al., 2017; Shim et al., 2001a, 2017) that is likely peridotitic or pyrolitic (McDonough and Sun, 1995).