View of the AP2/EREBP TF Family in Maize
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Document Downloaded From: This Paper Must Be Cited As: the Final
Document downloaded from: http://hdl.handle.net/10251/83362 This paper must be cited as: Fourquin ., C.; Ferrandiz Maestre, C. (2014). The essential role of NGATHA genes in style and stigma specification is widely conserved across eudicots. New Phytologist. 202(3):1001- 1013. doi:10.1111/nph.12703. The final publication is available at http://doi.org/10.1111/nph.12703 Copyright Wiley Additional Information 1 The essential role of NGATHA genes in style and stigma specification is widely 2 conserved across eudicots 3 Chloé Fourquin and Cristina Ferrándiz1 4 Instituto de Biología Molecular y Celular de Plantas, UPV-CSIC, 46022 Valencia, Spain 5 6 1 For correspondence : 7 Cristina Ferrándiz 8 Instituto de Biología Molecular y Celular de Plantas, UPV-CSIC, 46022 Valencia, Spain 9 Tel. +34 963877892; Fax +34 963877859; e-mail: [email protected] 10 11 Total word count for the main body of the text: 5721 12 13 Word count for individual sections are as follows: 14 • Introduction, 947 15 • Materials and Methods, 765 16 • Results, 2035 17 • Discussion, 1868 18 • Acknowledgements, 106 19 20 The manuscript contains 6 figures (5 in color) and 1 table. 21 Supplementary material consists of 4 figures and 1 table. 22 23 SUMMARY 24 • Carpel development and evolution is a central issue for plant biology. The 25 conservation of genetic functions conferring carpel identity has been widely studied 26 in higher plants. However, although genetic networks directing the development of 27 characteristic features of angiosperm carpels such as stigma and style are 28 increasingly known in Arabidopsis thaliana, little information is available on the 29 conservation and diversification of these networks in other species. -

Other Roles of Euap2 Genes in Plant Development Charles U
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 24 October 2019 doi:10.20944/preprints201901.0035.v3 Review Besides and Beyond Flowering: Other roles of euAP2 genes in plant development Charles U. Solomon1, 2* and Sinead Drea1 1Department of Genetics and Genome Biology, University of Leicester, UK; [email protected]; [email protected] 2Department of Plant Science and Biotechnology, Abia State University, Uturu, Nigeria *Correspondence: [email protected] Abstract: EuAP2 genes are famous for their role in flower development. A legacy of the founding member of this subfamily of transcription factors, whose mutants lacked petals in Arabidopsis. However, studies of euAP2 genes in several species have accumulated evidence highlighting the diverse roles of euAP2 genes in other aspects of plant development. Here, we emphasize other developmental roles of euAP2 genes in various species and suggest a shift from regarding euAP2 genes as just flowering genes to consider the global role they may be playing in plant development. We hypothesize that their almost universal expression profile and pleiotropic effects of their mutation suggest their involvement in fundamental plant development processes. Keywords: EuAP2 genes; Flowering; Plant Development Introduction APETALA 2 (AP2) genes are named after a series of Arabidopsis mutants characterized by homeotic transformations of their sepals to leaves and petals to staminoid petals. Analysis of the ap2 mutants along with other floral mutants gave birth to the ABC model of flower development where AP2 is classified as an A-class gene [1, 2]. The forerunner AP2 protein was cloned and characterized in Arabidopsis [3]. The Arabidopsis AP2 protein comprising 432 amino acids (aa) is mainly characterized by the possession of two AP2 domains, each made up of 68-aa with an 18-aa core conserved section that forms an amphipathic -훼–helix . -

Systems Biology Lecture Outline (Systems Biology) Michael K
Spring 2019 – Epigenetics and Systems Biology Lecture Outline (Systems Biology) Michael K. Skinner – Biol 476/576 CUE 418, 10:35-11:50 am, Tuesdays & Thursdays January 22 & 29, 2019 Weeks 3 and 4 Systems Biology (Components & Technology) Components (DNA, Expression, Cellular, Organ, Physiology, Organism, Differentiation, Development, Phenotype, Evolution) Technology (Genomics, Transcriptomes, Proteomics) (Interaction, Signaling, Metabolism) Omics (Data Processing and Resources) Required Reading ENCODE (2012) ENCODE Explained. Nature 489:52-55. Tavassoly I, Goldfarb J, Iyengar R. (2018) Essays Biochem. 62(4):487-500. Literature Sundaram V, Wang T. Transposable Element Mediated Innovation in Gene Regulatory Landscapes of Cells: Re-Visiting the "Gene-Battery" Model. Bioessays. 2018 40(1). doi: 10.1002/bies.201700155. Abil Z, Ellefson JW, Gollihar JD, Watkins E, Ellington AD. Compartmentalized partnered replication for the directed evolution of genetic parts and circuits. Nat Protoc. 2017 Dec;12(12):2493- 2512. Filipp FV. Crosstalk between epigenetics and metabolism-Yin and Yang of histone demethylases and methyltransferases in cancer. Brief Funct Genomics. 2017 Nov 1;16(6):320-325. Chen Z, Li S, Subramaniam S, Shyy JY, Chien S. Epigenetic Regulation: A New Frontier for Biomedical Engineers. Annu Rev Biomed Eng. 2017 Jun 21;19:195-219. Hollick JB. Paramutation and related phenomena in diverse species. Nat Rev Genet. 2017 Jan;18(1):5-23. Macovei A, Pagano A, Leonetti P, Carbonera D, Balestrazzi A, Araújo SS. Systems biology and genome-wide approaches to unveil the molecular players involved in the pre-germinative metabolism: implications on seed technology traits. Plant Cell Rep. 2017 May;36(5):669-688. Bagchi DN, Iyer VR. -

The Human Proteome Co-Regulation Map Reveals Functional Relationships Between Proteins
bioRxiv preprint doi: https://doi.org/10.1101/582247; this version posted March 19, 2019. The copyright holder for this preprint (which was 19/03/2019not certified by peer review) is the author/funder, who hasNBT_manuscript_revised granted bioRxiv a license - toGoogle display Docs the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 The human proteome co-regulation map reveals functional relationships 2 between proteins 3 Georg Kustatscher1 *, Piotr Grabowski2 *, Tina A. Schrader3 , Josiah B. Passmore3 , Michael 4 Schrader3 , Juri Rappsilber1,2# 5 1 Wellcome Centre for Cell Biology, University of Edinburgh, Edinburgh EH9 3BF, UK 6 2 Bioanalytics, Institute of Biotechnology, Technische Universität Berlin, 13355 Berlin, 7 Germany 8 3 Biosciences, University of Exeter, Exeter EX4 4QD, UK 9 * Equal contribution 10 # Communicating author: [email protected] 11 Submission as Resource article. 12 The annotation of protein function is a longstanding challenge of cell biology that 13 suffers from the sheer magnitude of the task. Here we present ProteomeHD, which 14 documents the response of 10,323 human proteins to 294 biological perturbations, 15 measured by isotope-labelling mass spectrometry. Using this data matrix and robust 16 machine learning we create a co-regulation map of the cell that reflects functional 17 associations between human proteins. The map identifies a functional context for 18 many uncharacterized proteins, including microproteins that are difficult to study with 19 traditional methods. Co-regulation also captures relationships between proteins 20 which do not physically interact or co-localize. For example, co-regulation of the 21 peroxisomal membrane protein PEX11β with mitochondrial respiration factors led us 22 to discover a novel organelle interface between peroxisomes and mitochondria in 23 mammalian cells. -

Comprehensive Genome-Wide Exploration of C2H2 Zinc Finger Family in Grapevine (Vitis Vinifera L.): Insights Into the Roles in the Pollen Development Regulation
G C A T T A C G G C A T genes Article Comprehensive Genome-Wide Exploration of C2H2 Zinc Finger Family in Grapevine (Vitis vinifera L.): Insights into the Roles in the Pollen Development Regulation Oscar Arrey-Salas 1,* , José Carlos Caris-Maldonado 2, Bairon Hernández-Rojas 3 and Enrique Gonzalez 1 1 Laboratorio de Genómica Funcional, Instituto de Ciencias Biológicas, Universidad de Talca, 3460000 Talca, Chile; [email protected] 2 Center for Research and Innovation (CRI), Viña Concha y Toro, Ruta k-650 km 10, 3550000 Pencahue, Chile; [email protected] 3 Ph.D Program in Sciences Mention in Modeling of Chemical and Biological Systems, Faculty of Engineering, University of Talca, Calle 1 Poniente, 1141, 3462227 Talca, Chile; [email protected] * Correspondence: [email protected] Abstract: Some C2H2 zinc-finger proteins (ZFP) transcription factors are involved in the development of pollen in plants. In grapevine (Vitis vinifera L.), it has been suggested that abnormalities in pollen development lead to the phenomenon called parthenocarpy that occurs in some varieties of this cultivar. At present, a network involving several transcription factors types has been revealed and key roles have been assigned to members of the C2H2 zinc-finger proteins (ZFP) family in model plants. However, particularities of the regulatory mechanisms controlling pollen formation in grapevine Citation: Arrey-Salas, O.; remain unknown. In order to gain insight into the participation of ZFPs in grapevine gametophyte Caris-Maldonado, J.C.; development, we performed a genome-wide identification and characterization of genes encoding Hernández-Rojas, B.; Gonzalez, E. ZFP (VviZFP family). -

Genome-Wide Analysis of the AP2/ERF Superfamily in Apple and Transcriptional Evidence of ERF Involvement in Scab Pathogenesis
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Open Archive Toulouse Archive Ouverte Open Archive Toulouse Archive Ouverte ( OATAO ) OATAO is an open access repository that collects the work of Toulouse researchers and makes it freely available over the web where possible. This is an author-deposited version published in: http://oatao.univ- toulouse.fr/ Eprints ID: 11480 To link to this article : DOI:10.1016/j.scienta.2012.12.017 URL: http://dx.doi.org/10.1016/j.scienta.2012.12.017 To cite this version: Girardi, Cesar Luis and Rombaldi, César Valmor and Dal Cero, Joceani and Nobile, Paula M. and Laurent, François and Bouzayen, Mondher and Quecini, Vera Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. (2013) Scientia Horticulturae, vol. 151. pp. 112-121. ISSN 0304-4238 Any correspondence concerning this service should be sent to the repository administrator: [email protected] ! Genome•wide analysis of the AP2 /ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis a b b c c César Luís Girardi , César Valmor Rombaldi , Joceani Dal Cero , Paula M. Nobile , Franc¸ ois Laurens , d a,∗ Mondher Bouzayen , Vera Quecini a Embrapa Uva e Vinho, Caixa Postal 130, 95700•000 Bento Gonc¸ alves, RS, Brazil b ◦ Universidade Federal de Pelotas, Faculdade de Agronomia Eliseu Maciel, Departamento de Ciência e Tecnologia Agroindustrial, s/n caixa postal 354, 96010•900 Pelotas, RS, Brazil c UMR 1259 Génétique et Horticulture (GenHort), Angers, France d UMR 990, Génomique et Biotechnologie des Fruits, INP, ENSAT, INRA, F•31320 Castanet Tolosan, France a b s t r a c t The APETALA2 (AP2)/ETHYLENE RESPONSE FACTOR (ERF) superfamily of transcriptional regulators is involved in several growth, development and stress responses processes in higher plants. -

Gene Family in Panax Ginseng CA Meyer
bioRxiv preprint doi: https://doi.org/10.1101/850164; this version posted November 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Structural variation, functional differentiation and 2 expression characteristics of the AP2/ERF gene family and 3 its response to cold stress and methyl jasmonate in Panax 4 ginseng C.A. Meyer 5 Short title: Structural and functional Analysis of AP2/ERF 6 gene family in Panax ginseng C.A. Meyer 1,2 1 1 1 1,2 1,2 7 Jing Chen ☯,Yuanhang Zhou ☯, Qi Zhang , Li Li , Chunyu Sun , Kangyu Wang , 8 Yanfang Wang2,3, Mingzhu Zhao1,2, Hongjie Li1, Yilai Han1, Ping Chen1, Ruiqi Li1, 9 Jun Lei1, Meiping Zhang 1,2*, and Yi Wang 1,2* 10 1 College of Life Science, Jilin Agricultural University, 2888 Xincheng Street, 11 Changchun, Jilin 130118, China. 12 2 Research Center for Ginseng Genetic Resources Development and Utilization, 13 2888 Xincheng Street, Changchun, Jilin 130118, China. 14 3 College of Chinese Medicinal Materials, Jilin Agricultural University, 2888 15 Xincheng Street, Changchun 130118, Jilin, China. 16 17 ☯These authors contributed equally to this work. 18 Correspondence should be addressed to [email protected] and 19 [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/850164; this version posted November 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

An Updated Overview on the Regulation of Seed Germination
An Updated Overview on the Regulation of Seed Germination Table S1. Selected recent reviews on seed germination-related aspects. Subject (Keywords) References Generalists ABA, Arabidopsis, dormancy, germination, GA, seed maturation. [3] GAs and ABA GAs, ABA, hormone signaling, seed germination, abiotic stresses, crosstalk of [10] hormone signaling. DELLAs, GAs, hormone crosstalk, phenotypic plasticity, plant development. [20] DELLA, GRAS, GAs, Arabidopsis, rice, GID1, crosstalk, [22] ABA, ABA synthesis, ABA signaling, ABA response. [35] PYR1, RCAR, ABA transduction, SnRK2, PP2C. [42] ABI5, abiotic stress response, phytohormone crosstalk, ABA, brassinosteroids, GAs, [81] jasmonates, posttranslational modification. Hormones transport GA biosynthesis, GA action, GA transport, evolution, Gibberella fujikuroi. [48] GA, plant hormone, hormone transport and localization, GA transporters, NPF, [49] SWEET. ABA; guard cell; transporter; water stress. [50] GA metabolism, GA transport, GA gradients, cellular growth. [51] Plant NRT1/PTR transporters, phylogenetic relationships, substrates. [72] Glucosinolate, hormone, nitrate, nitrite, peptide, transporter. [73] NIN-like protein (NLP), nitrate, nitrate-responsive gene expression, post- translational regulation, transcription factor, [77] nitrate signalling. Light regulation PIF signaling, Phytochromes, plant development. [109] Chromatin dynamics Shoot apical meristem, quiescent center, floral homeotic gene, polycomb repression, [162] seed development, dormancy, germination and greening. DNA methylation, -

Pollination-Induced Gene Changes That Lead to Senescence in Petunia × Hybrida
Pollination-Induced Gene Changes That Lead to Senescence in Petunia × hybrida DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Shaun Robert Broderick, M.S. Graduate Program in Horticulture and Crop Science The Ohio State University 2014 Dissertation Committee: Michelle L. Jones, Advisor Feng Qu Eric J. Stockinger Esther van der Knaap Copyrighted by Shaun Robert Broderick 2014 Abstract Flower longevity is a genetically programmed event that ends in flower senescence. Flowers can last from several hours to several months, based on flower type and environmental factors. For many flowers, particularly those that are ethylene- sensitive, longevity is greatly reduced after pollination. Cellular components are disassembled and nutrients are remobilized during senescence, which reduces the net energy expenditures of floral structures. The goal of this research is to identify the genes that can be targeted to extent shelf life by inhibiting pollination-induced senescence. Identifying and characterizing regulatory shelf-life genes will enable breeders to incorporate specific alleles that improve post production quality into ethylene-sensitive crops. Petunia × hybrida is particularly amenable to flower longevity studies because of its large floral organs, predictable flower senescence timing, and importance in the greenhouse industry. A general approach to gene functional analysis involves reducing gene expression and observing the resulting phenotype. Viruses, such as tobacco rattle virus (TRV), can be used to induce gene silencing in plants like petunia. We optimized several parameters that improved virus-induced gene silencing (VIGS) in petunia by increasing the consistency and efficiency of silencing. -

A Latent Ability to Persist: Differentiation in Toxoplasma Gondii
View metadata, citation and similar papers at core.ac.uk brought to you by CORE HHS Public Access provided by IUPUIScholarWorks Author manuscript Author ManuscriptAuthor Manuscript Author Cell Mol Manuscript Author Life Sci. Author Manuscript Author manuscript; available in PMC 2019 July 01. Published in final edited form as: Cell Mol Life Sci. 2018 July ; 75(13): 2355–2373. doi:10.1007/s00018-018-2808-x. A Latent Ability to Persist: Differentiation in Toxoplasma gondii Victoria Jeffers1,*, Zoi Tampaki2, Kami Kim2,3, and William J. Sullivan Jr1,4 1Department of Pharmacology and Toxicology, Indiana University School of Medicine, Indianapolis, IN 46202, USA 2Departments of Medicine, Microbiology & Immunology, and Pathology; Albert Einstein College of Medicine, Bronx, NY 10461, USA 3Department of Internal Medicine, Morsani College of Medicine, University of South Florida, Tampa, FL 33612, USA 4Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN 46202, USA Abstract A critical factor in the transmission and pathogenesis of Toxoplasma gondii is the ability to convert from an acute disease-causing, proliferative stage (tachyzoite), to a chronic, dormant stage (bradyzoite). The conversion of the tachyzoite-containing parasitophorous vacuole membrane into the less permeable bradyzoite cyst wall, allows the parasite to persist for years within the host to maximize transmissibility to both primary (felids) and secondary (virtually all other warm-blooded vertebrates) hosts. This review presents our current understanding of the latent stage, including the factors that are important in bradyzoite induction and maintenance. Also discussed are the recent studies that have begun to unravel the mechanisms behind stage switching. Keywords Toxoplasma; toxoplasmosis; differentiation; encystation; tachyzoite; bradyzoite; latency; gene regulation; epigenetics; immunity INTRODUCTION Toxoplasma gondii is an obligate intracellular protozoan parasite capable of infecting warm- blooded vertebrates world-wide [1]. -

Roles of Wrky Proteins in Mediating the Crosstalk of Hormone Signaling Pathways: an Approach Integrating Bioinformatics and Experimental Biology
UNLV Retrospective Theses & Dissertations 1-1-2006 Roles of Wrky proteins in mediating the crosstalk of hormone signaling pathways: An approach integrating bioinformatics and experimental biology Zhen Xie University of Nevada, Las Vegas Follow this and additional works at: https://digitalscholarship.unlv.edu/rtds Repository Citation Xie, Zhen, "Roles of Wrky proteins in mediating the crosstalk of hormone signaling pathways: An approach integrating bioinformatics and experimental biology" (2006). UNLV Retrospective Theses & Dissertations. 2711. http://dx.doi.org/10.25669/0cw1-sipg This Dissertation is protected by copyright and/or related rights. It has been brought to you by Digital Scholarship@UNLV with permission from the rights-holder(s). You are free to use this Dissertation in any way that is permitted by the copyright and related rights legislation that applies to your use. For other uses you need to obtain permission from the rights-holder(s) directly, unless additional rights are indicated by a Creative Commons license in the record and/or on the work itself. This Dissertation has been accepted for inclusion in UNLV Retrospective Theses & Dissertations by an authorized administrator of Digital Scholarship@UNLV. For more information, please contact [email protected]. ROLES OF WRKY PROTEINS IN MEDIATING THE CROSSTALK OF HORMONE SIGNALING PATHWAYS: AN APPROACH INTEGRATING BIOINFORMATICS AND EXPERIMENTAL BIOLOGY by Zhen Xie Bachelor of Sciences Shandong Agricultural University 1998 Master of Sciences Shandong Agricultural University 2001 A dissertation submitted in partial fulfillment of the requirements for the Doctor of Philosophy Degree in Biological Sciences Department of Biological Sciences College of Sciences Graduate College University of Nevada, Las Vegas December 2006 Reproduced with permission of the copyright owner. -
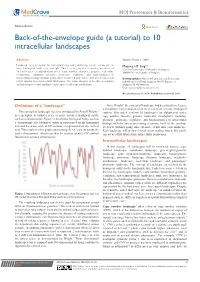
Back-Of-The-Envelope Guide (A Tutorial) to 10 Intracellular Landscapes
MOJ Proteomics & Bioinformatics Review Article Open Access Back-of-the-envelope guide (a tutorial) to 10 intracellular landscapes Abstract Volume 7 Issue 1 - 2018 Landscape is a metaphor for conceptualizing and visualizing a score across one or Maurice HT Ling1,2 more biological entities or concepts. This review provides a cursory overview of 1Colossus Technologies LLP, Republic of Singapore 10 landscapes (in alphabetical order, copy number, fluxome, genome, molecular, 2HOHY Pte. Ltd., Republic of Singapore metabolome, mutation, phenome, proteome, regulome, and transcriptome) in intracellular biology without going into extensive depth; hence, this article can act as Correspondence: Maurice HT Ling, Colossus Technologies a first tutorial into intracellular landscapes. The value ahead is to be able to compare LLP, 8 Burns Road, Trivex, Singapore 369977, Republic of and interrogate across multiple landscapes at different resolutions. Singapore, Tel +6596669233, Email [email protected] Received: January 29, 2018 | Published: February 06, 2018 Definition of a “landscape” Since Wright,1 the concept of landscape had been used to reference a metaphoric representation of an overview of one or more biological 1 The concept of landscape was first introduced by Sewell Wright, entities. This article reviews 10 landscapes (in alphabetical order, as a metaphor to visual a scale or score across a biological entity, copy number, fluxome, genome, molecular, metabolome, mutation, such as a chromosome (Figure 1). In a linear biological entity, such as phenome, proteome, regulome, and transcriptome) in intracellular a chromosome; the biological entity is represented on the horizontal biology with the aim of presenting a cursory, back of the envelope axis and the score, such as GC content, is represented on the vertical overview without going into extensive depth into each landscape.