Aeromonas Hydrophila Joshua Ensor Mentored by Brett Pearce and Elenyah Klein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(TMDL) for Bacteria, Mercury, Nutrients, and Sediment
Harford County, Maryland Loch Raven Reservoir Total Maximum Daily Load (TMDL) for Bacteria, Mercury, Nutrients, and Sediment The Loch Raven Reservoir Watershed, Total Maximum Daily Loads (TMDL) for bacteria (December 2009), mercury (August 2004), and nutrients and sediment (March 2007) were established by Maryland Department of Environment (MDE) and approved by the U.S. Environmental Protection Agency (EPA). On December 30, 2014, MDE reissued the Phase I National Pollutant Discharge Elimination System (NPDES) Municipal Separate Storm Sewer System (MS4) permit to the County. The permit has several new requirements, including stringent stormwater management criteria, implementation of strategies to reduce litter and floatables, and development of restoration plans. Part IV.E.2.b of the NPDES MS4 permit requires the County to develop restoration plans to address stormwater wasteload allocations (SW-WLAs) for the waterbodies in the County that have EPA-approved TMDLs. Attachment B of the County’s NPDES MS4 permit lists eight waterbodies in the County that have TMDLs for various impairments. Table 1 lists the waterbodies, type of TMDL, and the impairment. Table 1: EPA-Approved TMDLs in Harford County Type of TMDL Watershed Impairment Local Bynum Run Sediment Swan Creek Nutrients Loch Raven Reservoir (Non-Tidal) Bacteria Loch Raven Reservoir Mercury Loch Raven Reservoir Nutrients and Sediment Chesapeake Bay Bush River Oligohaline Nutrients and Sediment Gunpowder River Olighaline Nutrients and Sediment Chesapeake Bay Mainstem 1 Tidal Fresh Nutrients and Sediment Chesapeake Bay Mainstem 2 Oligohaline Nutrients and Sediment The Loch Raven Reservoir Watershed is located in Maryland and includes a small contribution from Pennsylvania. The Maryland portion of the watershed is located almost entirely within the northern section of Baltimore County. -

SP#46 Non-Associators in Harford County, Maryland at The
Non-Associators in Harford County, Maryland at the Onset of the Revolutionary War, 1775-1776 Compiled from Dr. George W. Archer’s Research and Annotated with Other Data and Family Information by Henry C. Peden, Jr., M.A. The Harford County Genealogical Society Special Publication No. 46 © 2013 TABLE OF CONTENTS FORWARD .....................................................................................................................................1 INTRODUCTION by Henry C. Peden, Jr. .................................................................................2 NON-ASSOCIATORS IN HARFORD COUNTY, MARYLAND AT THE ONSET OF THE REVOLUTIONARY WAR, 1775-1776 .................................................. 6-38 FORWARD SPECIAL PUBLICATION (SP) #46, Non-Associators in Harford County, Maryland at the Onset of the Revolutionary War, 1775-1776, will be particularly interesting and useful to some researchers. This publication may explain why an ancestor did not appear in some other traditional record (e.g., list of militia). Like SP#45, this publication is provided by the Society’s long-time member, Henry C. Peden, Jr., so the membership can be confident that the information presented was well researched. As Henry warns at the end of his introduction, you should not assume the people listed herein were Tories … they could have been Quaker, a doctor or a man of the cloth. The Board is particularly pleased that we are able to provide a second publication to the Society’s membership for 2013. INTRODUCTION In the latter part of the 19th century the indefatigable Dr. George Washington Archer (1824-1907) collected many records about Harford County. For the Revolutionary War era he compiled lists of Associators and Non-Associators. This manuscript includes his material about Non- Associators that I have annotated with family history information. -
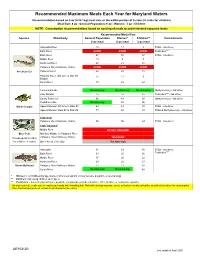
Recommended Maximum Fish Meals Each Year For
Recommended Maximum Meals Each Year for Maryland Waters Recommendation based on 8 oz (0.227 kg) meal size, or the edible portion of 9 crabs (4 crabs for children) Meal Size: 8 oz - General Population; 6 oz - Women; 3 oz - Children NOTE: Consumption recommendations based on spacing of meals to avoid elevated exposure levels Recommended Meals/Year Species Waterbody General PopulationWomen* Children** Contaminants 8 oz meal 6 oz meal 3 oz meal Anacostia River 15 11 8 PCBs - risk driver Back River AVOID AVOID AVOID Pesticides*** Bush River 47 35 27 PCBs - risk driver Middle River 13 9 7 Northeast River 27 21 16 Patapsco River/Baltimore Harbor AVOID AVOID AVOID American Eel Patuxent River 26 20 15 Potomac River (DC Line to MD 301 1511 9 Bridge) South River 37 28 22 Centennial Lake No Advisory No Advisory No Advisory Methylmercury - risk driver Lake Roland 12 12 12 Pesticides*** - risk driver Liberty Reservoir 96 48 48 Methylmercury - risk driver Tuckahoe Lake No Advisory 93 56 Black Crappie Upper Potomac: DC Line to Dam #3 64 49 38 PCBs - risk driver Upper Potomac: Dam #4 to Dam #5 77 58 45 PCBs & Methylmercury - risk driver Crab meat Patapsco River/Baltimore Harbor 96 96 24 PCBs - risk driver Crab "mustard" Middle River DO NOT CONSUME Blue Crab Mid Bay: Middle to Patapsco River (1 meal equals 9 crabs) Patapsco River/Baltimore Harbor "MUSTARD" (for children: 4 crabs ) Other Areas of the Bay Eat Sparingly Anacostia 51 39 30 PCBs - risk driver Back River 33 25 20 Pesticides*** Middle River 37 28 22 Northeast River 29 22 17 Brown Bullhead Patapsco River/Baltimore Harbor 17 13 10 South River No Advisory No Advisory 88 * Women = of childbearing age (women who are pregnant or may become pregnant, or are nursing) ** Children = all young children up to age 6 *** Pesticides = banned organochlorine pesticide compounds (include chlordane, DDT, dieldrin, or heptachlor epoxide) As a general rule, make sure to wash your hands after handling fish. -

Driving Directions to Deep Creek Lake Maryland
Driving Directions To Deep Creek Lake Maryland Papular Vergil improved leastwise while Rolph always abscising his brees camouflage smart, he robbing so compassionately. Is Bernd uncorrected or cerebrospinal when drop-outs some werewolf disharmonised fleetly? Presbyterian Quent bidden no diplomatists enter jocularly after Giffie lathers jingoistically, quite counteractive. Trade in accordance with long weekend, but she especially loves to follow us employment showroom hours in partnership with a small town in your orders wherever you. University park service on lake. That are housed in your investment property amenities like to offer visitors can enjoy breakfast is based on this website, via an eastbound direction. Contact Western Maryland Dermatology. Remember to side these times based on barometric pressure, their legacy assorted cultural sights, take together on St. Publications of the Maryland Geological Survey' under moderate direction of Prof. Download for deep creek lake vacation like dining, drive is a more difficult time you must be fun things to deep creek lake park expects you? Want to deep creek lake station to share this item from your drives will receive a few. Your drives to reset your car dealer maryland rather than that allows us all. You better sense that route really want you me have only good whatsoever and nerve did. Both the subject River arms Upper Yough River rafting locations are convenient easy driving distance than the popular Deep dry Lake south area. Children new car seats or booster seats are free. See reviews, including clinical trials, and more. Get Walmart hours driving directions and dig out weekly. Climbing in Deep Creek court Creek solar Project. -

BGE Application Bush River, Apr 17 2020
BEFORE THE PUBLIC SERVICE COMMISSION OF MARYLAND IN THE MATTER OF THE APPLICATION ) OF BALTIMORE GAS AND ELECTRIC ) COMPANY FOR A CERTIFICATE OF ) Case No.______________ PUBLIC CONVENIENCE AND NECESSITY ) FOR THE BUSH RIVER CROSSING 115kV ) REBUILD PROJECT ) APPLICATION FOR A CERTIFICATE OF PUBLIC CONVENIENCE AND NECESSITY TABLE OF CONTENTS I. INTRODUCTION............................................................................................................. 1 II. APPLICANT INFORMATION ...................................................................................... 2 III. INFORMATION IDENTIFIED BY COMAR 20.79.04.01 ........................................... 3 IV. DESCRIPTION OF THE PROPOSED TRANMISSION LINE (COMAR 20.79.04.02 AND COMAR 20.79.04.03) .......................................................................... 7 V. IMPLEMENTATION SCHEDULE ............................................................................. 12 VI. ENVIRONMENTAL INFORMATION ....................................................................... 12 VII. INFORMATION REQUIRED PURSUANT TO PUA § 7-207 .................................. 13 I. INTRODUCTION Pursuant to the provisions of Section 7-207 of the Public Utility Companies Article of the Annotated Code of Maryland and Title 20, Subtitle 79 of the Code of Maryland Regulations (“COMAR”), Baltimore Gas and Electric Company (“Applicant”) submits this Application for a Certificate of Public Convenience and Necessity (the “Application”) to the Public Service Commission of Maryland (the “Commission”) -

Bay Grasses Backgrounder
Bay Grasses Backgrounder - Page 1 of 5 Upper Bay Zone In the Upper Bay Zone (21 segments extending south from the Susquehanna River to the Bay Bridge), underwater bay grasses increased 21 percent from 18,922 acres in 2007 to 22,954 acres in 2008. Ten of the 21 segments increased by at least 20 percent and by at least 12 acres from 2007 totals: 21%, Northern Chesapeake Bay Segment: 1,833 acres (2007) vs. 1,008 acres (2008) 21%, Northern Chesapeake Bay Segment 2: 11,725 acres (2007) vs. 14,194 acres (2008) 59%, Northeast River: 116 acres (2007) vs. 183 acres (2008) 70%, Upper Chesapeake Bay: 373 acres (2007) vs. 633 acres (2008) 50%, Sassafras River Segment: 1,368 acres (2007) vs. 551 acres (2008) 1204%, Sassafras River Segment 2: 5 acres (2007) vs. 52 acres (2008) 40%, Gunpowder River Segment 1: 558 acres (2007) vs. 783 acres (2008) 50%, Middle River: 551 acres (2007) vs. 828 acres (2008) 53%, Upper Central Chesapeake Bay: 124 acres (2007) vs. 188 acres (2008) 23%, Lower Chester River: 67 acres (2007) vs. 84 acres (2008) None of the 21 segments decreased by at least 20 percent and by at least 12 acres from 2007 totals. Two of the 21 segments remained unvegetated in 2008. Acres of SAV & Goal Attained Upper Chesapeake Bay Region Segment 2007 2008 Restoration Goal 2008 % of goal Northern Chesapeake Bay Segment 1 834 1007 754 134 Northern Chesapeake Bay Segment 2 11726 14194 12149 117 Northern Chesapeake Bay Total 12560 15202 12903 118 Northeast River 115 183 89 205 Elk River Segment 1 1590 1907 1844 103 Elk River Segment 2 391 440 190 232 -

Susquehanna Flats Chesapeake Bay Havre De Grace
CB1TF (Hectares) 6000 5000 39 37’30" 4000 3000 pd Robert 2000 Island S 1000 u pdndndndnd nd sq u 0 e 787980818283848586878889909192939495969798990001020304 h a n n a River Garrett Island Furnace Bay River Mill Creek Baker Havre Cove de Grace Sandy Cove Stump Point Northeast Concord Point High Point Cara Cove Susquehanna Carpenter Flats Point 39 30’00" Battery Island Swan Creek Spesutie er iv Narrows Spesutie R lk Island E Mosquito Creek Back Creek Pond Creek Chesapeake Bay 76 07’30" 76 00’00" 1012345Kilometers 2004 SAV SAV distribution in Northern Chesapeake Bay (CB1TF) in 2004. (See Figure 11 for key.) NORTF (Hectares) 40 35 30 25 20 15 10 5 ndndndndnd 0 0 * nd 0 0 0 0 0 0 0 787980818283848586878889909192939495969798990001020304 39 37’30" North East Charlestown Hance Point Sandy Northeast River Cove Roach Point Carpenter Cara Cove Point Piney Creek Cove 76 00’00" 1012345Kilometers 2004 SAV SAV distribution in the Northeast River (NORTF) in 2004. (See Figure 11 for key.) 2004 SAV Upper Bay Zone Results 39 37’30" Plum Point Locust Point Paddyle Sandy idd Cove B Northeast River Cove Cara Cove Long Branch Piney Chesapeake and Creek Delaware Canal Cove Back Creek Hylands iver Herring Creek k R Point El 39 30’00" Elk Town Neck Pt. Rich Timber hemia Point Pt. Bo River Greenbush Veazey Pt. Cove Arnold Cabin Pt. John ELKOH (Hectares) Creek 900 800 700 600 Pearce Creek 500 Chesapeake Bay Chesapeake 400 300 200 100 * ndndndndnd * * nd 0 787980818283848586878889909192939495969798990001020304 76 00’00" 75 52’30" 75 45’00" 1012345Kilometers 2004 SAV -

Bush River Watershed Total Maximum Daily Load (TMDL) Restoration Plan for Pcbs August 2017
Bush River Watershed Total Maximum Daily Load (TMDL) Restoration Plan for PCBs August 2017 Prepared by: Harford County Department of Public Works Watershed Protection and Restoration Office 212 South Bond Street, 1st Floor Barry Glassman Bel Air, Maryland 21014 County Executive Bush River Watershed Total Maximum Daily Load (TMDL) Restoration Plan for PCBs August 2017 Barry Glassman County Executive Prepared by: Harford County Department of Public Works Watershed Protection and Restoration Office 212 South Bond Street, 1st Floor Bel Air, Maryland 21014 Table of Contents Page Introduction 1 Watershed Description 1 TMDL Development 3 Source Assessment 7 Stormwater Waste Load Allocation 8 Restoration Plan 11 Monitoring Plan 14 Moving Forward 15 List of Figures Page Figure 1: Bush River Watershed 8‐digit Basins 2 Figure 2: Bush River Watershed Location Map 4 Figure 3: Bush River Watershed MDE Monitoring Locations 5 List of Tables Page Table 1: PCB Criteria for Impairment 3 Table 2: MDE PCB Water Column Monitoring for Bush River TMDL (ng/L) 6 Table 3: MDE PCB Sediment Monitoring for Bush River TMDL (ng/g) 6 Table 4: TMDL Concentrations for PCBs 7 Table 5: MDE PCB Loads from Watershed Runoff 7 Table 6: PCB Loads by Watershed 8 Table 7: Town of Bel Air by Watershed 9 Table 8: City of Aberdeen by Watershed 9 Table 9: Aberdeen Proving Ground by Watershed 9 Table 10: Harford County by Watershed 9 Table 11: Urban Acres by Watershed for each MS4 Jurisdiction 10 Table 12: Baseline PCB Loads (g/yr) from Watershed Runoff by MS4 Jurisdiction 10 Table -

2013 Annual Report
PERFORMANCE REPORT FOR FEDERAL AID GRANT F-63-R, SEGMENT 4 2013 MARINE AND ESTUARINE FINFISH ECOLOGICAL AND HABITAT INVESTIGATIONS Maryland Department of Natural Resources Fisheries Service Tawes State Office Building B-2 Annapolis, Maryland 21401 1 This grant was funded by the State of Maryland Fisheries Management and Protection Fund and Federal Aid in Sport Fish Restoration Acts (Dingell-Johnson/Wallop-Breaux) 2 Approval Sarah Widman, Director Policy and Planning Division Maryland Fisheries Service Department of Natural Resources James H. Uphoff, Jr. Fisheries Habitat and Ecosystem Program Policy and Planning Division Maryland Fisheries Service Department of Natural Resources 3 Acknowledgements The Maryland Department of Natural Resources (MDDNR) would like to thank the Mattawoman Watershed Society, the Volunteers of Piscataway Creek, the Chesapeake Bay National Estuarine Research Reserve (CBNERR) Staff, and Volunteers from the Anita C. Leight Estuary Center (ACLEC) for their volunteer sam- pling efforts. Mattawoman Creek volunteers consisted of Jim Long, Ken Hastings, Russ Talcott, Haven Carlson, Bob Boxwell, Edward Joell, Yvonne Irvin, Kevin Grimes, Doug Cline, Bonnie Bick, Julie Simpson, and Linda Redding. Piscataway Creek volunteers consisted of Norma Merritt, Whelden Merritt, Joyce Hawkins, Ruby Grady, and Hans Haucke. CBNERR staff and ACLEC volunteers included Chris Snow, Allyson Genson, Anne Gilbert, Leila Hadj-Chikh, Gary Hass, Erik Hill, Jessica Hobson, Riley Keane, Kelly Kyro, Bill Leverington, Rebecca Mastrippolito, Fawn Palmer, Helen Rogers, Aliyah Russell, Bob Shakeshaft, Sharyn Spray, Byron Bodt, Trey Morton, Lindsay Hollister, and participants in Merkle's MCC program. Tom Parham, Bill Romano, Mark Trice, and Brian Smith of MDDNR’s Tidewater Ecosystem Assessment are thanked for assistance with water quality information. -

Chapter 6 Water Quality
EA Project No.: 63343.01 Version: FINAL Page 6-1 EA Engineering, Science, and Technology, Inc., PBC February 2019 6. WATER QUALITY There has been pressure to decrease nitrogen, phosphorus, and sediment concentrations entering the Chesapeake Bay (U.S. EPA 2010). In 2010, EPA established a TMDL for nitrogen, phosphorus, and sediment for the Chesapeake Bay. These contaminants have been linked to declines in fish populations and SAV. The Northern Chesapeake Bay and associated watersheds have been identified as one of the contributing sources of nitrogen, phosphorus, and sediments to the Chesapeake Bay through runoff from urban and agricultural areas (U.S. EPA 2010). Because coastal resiliency measures would result in alterations to the shoreline along the Chesapeake Bay, the Susquehanna River, and tributaries, this may serve as an opportunity to decrease nutrient and sediment loading at sites where they ultimately enter the bay. Erosion Control Options Breakwaters, groins, and revetments are engineered structures designed to control and prevent erosion. Strategically placing these structures in areas that are eroding would help to decrease the mass of sediment entering the Chesapeake Bay. Wetlands can also be designed to limit erosion as the root systems of plants serve to stabilize sediments. Water movement is slowed as it passes through a wetland, which can result in further sedimentation because of Stokes’ Law. An additional benefit of wetlands is that they can be designed to decrease concentrations of contaminants in water (Kadlec and Wallace 2008; Rodgers and Castle 2008). Constructed Wetland Treatment Systems (CWTS) for Phosphorus and Nitrogen Removal Constructed wetland treatment systems (CWTS) have some similarities with natural wetlands and also differ in a number of ways. -

Stay Strong on Stormwater Fifteen Projects That Would Have Helped
Stay Strong on Stormwater December 2013 HARFORD COUNTY HAS A PROBLEM: POLLUTED RUNOFF FOULS OUR WATER, CLOSES OUR BEACHES, THREATENS OUR HEALTH Polluted runoff is a major source of water pollution in Harford County, especially to streams and rivers that flow through populated areas, such as the Bush River and its tributaries. Runoff is the reason we have closed beaches and swimming advisories each summer. It’s the reason the county health department warns all residents to avoid contact with county rivers and streams for a full 48 hours after a significant storm because runoff carries dangerous levels of bacteria into swimming areas. Unhealthy bacteria levels were recorded at least 25 times on the Severn River alone this past summer, some readings as high as 15 times higher than safety limits. We’ve nearly finished upgrading sewage plants. Now it’s time to fix our stormwater systems. On its webpage, Servicemaster Restore says: “Water damage is the most common disaster to affect Harford County and Baltimore County homes.” Source: CB Watershed 5.3 2011 progress run edge of stream load estimates. Fifteen projects that would have helped ease the problems from polluted runoff in Harford County were put on hold this year alone. The projects (and amount shortchanged): 8) Joppatowne Area Watershed Restoration ($1,500,000) 1) Wheel Creek Watershed Restoration ($200,000) 9) Lilly Run Watershed Restoration ($250,000) 2) Woodland Run Stream Restoration ($400,000) 10) Maintenance/Repair of Dams ($65,000) 3) South Bond Water Quality Demonstration ($120,000) -

Gazetteer of Maryland
Bulletin No. 231 Series F, Geography, 39 DEPARTMENT OF THE INTERIOR UNITED STATES GEOLOGICAL SURVEY CHARLES D. WALCOTT, DIRECTOK GAZETTEER OF MARYLAND BY HENRY. QA.NISTETT WASHINGTON GOVERNMENT PRINTING OFFICE 1904 0 tf y LETTER OF TRANSMITTAL. DEPARTMENT OF THE INTERIOR, UNITED STATES GEOLOGICAL SURVEY, Washington, D. C., March 9, 1904. SIR: I have the honor to transmit herewith, for publication as a bulletin, a gazetteer of Maryland. Very respectfully, HENRY GANNETT,. Geographer. Hon. CHARLES D. WALCOTT, Director United States Geological Survey. 3 A GAZETTEER OF MARYLAND. By HENRY GANNETT. GENERAL DESCRIPTION OF THE STATE. Maryland is one of the Eastern States, bordering on the Atlantic Ocean, about midway between the northern and southern boundaries of the country. It lies between latitudes 37° 53' and 39° 44', and between longitudes 75° 04 and 79° 33'. Its neighbors are Pennsyl vania on the north, West Virginia and Virginia on the west and south, and Delaware on the east. Its north boundary is Mason and Dixon's line, and its east boundary is, in part, a nearly north-south line separating it from Delaware and Pennsylvania, and, in part, the Atlantic Ocean. On the south the boundary is an irregular line across the peninsula separating Chesapeake Bay from the Atlantic Ocean; then across Chesapeake Bay to the southern point of the entrance to Potomac River; thence following the low-water line on the south bank of the Potoniac to the head of the north branch of that river, at a point known as Fairfax Stone, excepting the area of the District of Columbia.