Work Package 5 - a Genetically-Based Strategy to Identify New Targets in Ldl- C Metabolism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ABSTRACT ANGSTADT, ANDREA Y. Evaluation of the Genomic
ABSTRACT ANGSTADT, ANDREA Y. Evaluation of the Genomic Aberrations in Canine Osteosarcoma and Their Resemblance to the Human Counterpart. (Under the direction of Dr. Matthew Breen). In the last decade the domestic dog has emerged as an ideal biomedical model of complex genetic diseases such as cancers. Cancer in the dog occurs spontaneously and several studies have concluded that human and canine cancers have similar characteristics such as presentation of disease, rate of metastases, genetic dysregulation, and survival rates. Furthermore, in the genomic era the dog genome was found more homologous in sequence conservation to humans than mice, making it a valuable model organism for genetic study in addition to pathophysiological analysis. Osteosarcoma (OS), the most commonly diagnosed malignant bone tumor in humans and dogs, is one such cancer that would benefit from comparative genomic analysis. In humans, OS is a rare cancer diagnosed in fewer than 1,000 people per year in the USA, while in the domestic dog population the annual number of new cases is estimated to far exceed 10,000. This high rate of disease occurrence in dogs provides a unique opportunity to study the genomic imbalances in canine OS and their translational value to human OS as a means to identify important alterations involved in disease etiology. OS in humans is characterized by extremely complex karyotypes which contain both structural changes (translocations and/or rearrangements) and DNA copy number changes. Metaphase and array comparative genomic hybridization (aCGH) has assisted in uncovering the genetic imbalances that are associated with human OS phenotype. In dog OS, previous low-resolution (10-20Mb) aCGH analysis identified a wide range of recurrent copy number aberrations (CNAs), indicative of a similar level of genomic instability to human OS. -

PRODUCT SPECIFICATION Anti-C12orf43
Anti-C12orf43 Product Datasheet Polyclonal Antibody PRODUCT SPECIFICATION Product Name Anti-C12orf43 Product Number HPA046148 Gene Description chromosome 12 open reading frame 43 Clonality Polyclonal Isotype IgG Host Rabbit Antigen Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: AWGLEQRPHVAGKPRAGAANSQLSTSQPSLRHKVNEHEQDGNELQTTPEF RAHVAKKLGALLDSFITISEAAKEPAKAKVQKVALEDDGFRLFFTSVPGG REKEESPQPR Purification Method Affinity purified using the PrEST antigen as affinity ligand Verified Species Human Reactivity Recommended IHC (Immunohistochemistry) Applications - Antibody dilution: 1:50 - 1:200 - Retrieval method: HIER pH6 WB (Western Blot) - Working concentration: 0.04-0.4 µg/ml ICC-IF (Immunofluorescence) - Fixation/Permeabilization: PFA/Triton X-100 - Working concentration: 0.25-2 µg/ml Characterization Data Available at atlasantibodies.com/products/HPA046148 Buffer 40% glycerol and PBS (pH 7.2). 0.02% sodium azide is added as preservative. Concentration Lot dependent Storage Store at +4°C for short term storage. Long time storage is recommended at -20°C. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. For protocols, additional product information, such as images and references, see atlasantibodies.com. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. -

Mutations in Diphosphoinositol- Pentakisphosphate Kinase PPIP5K2 Are Associated with Hearing Loss in Human and Mouse
RESEARCH ARTICLE Mutations in Diphosphoinositol- Pentakisphosphate Kinase PPIP5K2 are associated with hearing loss in human and mouse Rizwan Yousaf1, Chunfang Gu2, Zubair M. Ahmed3, Shaheen N. Khan4, Thomas B. Friedman5, Sheikh Riazuddin6, Stephen B. Shears2, Saima Riazuddin1* a1111111111 1 Laboratory of Molecular Genetics, Department of Otorhinolaryngology-Head & Neck Surgery, School of Medicine University of Maryland, Baltimore, MD, United States of America, 2 Signal Transduction Laboratory, a1111111111 National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, a1111111111 NC, United States of America, 3 Laboratory of Neurogenetics and Regenerative Medicine, Department of a1111111111 Otorhinolaryngology-Head & Neck Surgery, School of Medicine University of Maryland, Baltimore, MD, United a1111111111 States of America, 4 National Center for Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan, 5 Section on Human Genetics, Laboratory of Molecular Genetics, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, MD, United States of America, 6 Shaheed Zulfiqar Ali Bhutto Medical University, Pakistan Institute of Medical Sciences, Islamabad, Pakistan * [email protected] OPEN ACCESS Citation: Yousaf R, Gu C, Ahmed ZM, Khan SN, Friedman TB, Riazuddin S, et al. (2018) Mutations Abstract in Diphosphoinositol-Pentakisphosphate Kinase PPIP5K2 are associated with hearing loss in Autosomal recessive nonsyndromic hearing loss is a genetically heterogeneous disorder. human and mouse. PLoS Genet 14(3): e1007297. https://doi.org/10.1371/journal.pgen.1007297 Here, we report a severe-to-profound sensorineural hearing loss locus, DFNB100 on chro- mosome 5q13.2-q23.2. Exome enrichment followed by massive parallel sequencing Editor: Karen B. Avraham, Tel Aviv University, ISRAEL revealed a c.2510G>A transition variant in PPIP5K2 that segregated with DFNB100-associ- ated hearing loss in two large apparently unrelated Pakistani families. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Evidence for Differential Alternative Splicing in Blood of Young Boys With
Stamova et al. Molecular Autism 2013, 4:30 http://www.molecularautism.com/content/4/1/30 RESEARCH Open Access Evidence for differential alternative splicing in blood of young boys with autism spectrum disorders Boryana S Stamova1,2,5*, Yingfang Tian1,2,4, Christine W Nordahl1,3, Mark D Shen1,3, Sally Rogers1,3, David G Amaral1,3 and Frank R Sharp1,2 Abstract Background: Since RNA expression differences have been reported in autism spectrum disorder (ASD) for blood and brain, and differential alternative splicing (DAS) has been reported in ASD brains, we determined if there was DAS in blood mRNA of ASD subjects compared to typically developing (TD) controls, as well as in ASD subgroups related to cerebral volume. Methods: RNA from blood was processed on whole genome exon arrays for 2-4–year-old ASD and TD boys. An ANCOVA with age and batch as covariates was used to predict DAS for ALL ASD (n=30), ASD with normal total cerebral volumes (NTCV), and ASD with large total cerebral volumes (LTCV) compared to TD controls (n=20). Results: A total of 53 genes were predicted to have DAS for ALL ASD versus TD, 169 genes for ASD_NTCV versus TD, 1 gene for ASD_LTCV versus TD, and 27 genes for ASD_LTCV versus ASD_NTCV. These differences were significant at P <0.05 after false discovery rate corrections for multiple comparisons (FDR <5% false positives). A number of the genes predicted to have DAS in ASD are known to regulate DAS (SFPQ, SRPK1, SRSF11, SRSF2IP, FUS, LSM14A). In addition, a number of genes with predicted DAS are involved in pathways implicated in previous ASD studies, such as ROS monocyte/macrophage, Natural Killer Cell, mTOR, and NGF signaling. -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Methods in and Applications of the Sequencing of Short Non-Coding Rnas" (2013)
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2013 Methods in and Applications of the Sequencing of Short Non- Coding RNAs Paul Ryvkin University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Bioinformatics Commons, Genetics Commons, and the Molecular Biology Commons Recommended Citation Ryvkin, Paul, "Methods in and Applications of the Sequencing of Short Non-Coding RNAs" (2013). Publicly Accessible Penn Dissertations. 922. https://repository.upenn.edu/edissertations/922 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/922 For more information, please contact [email protected]. Methods in and Applications of the Sequencing of Short Non-Coding RNAs Abstract Short non-coding RNAs are important for all domains of life. With the advent of modern molecular biology their applicability to medicine has become apparent in settings ranging from diagonistic biomarkers to therapeutics and fields angingr from oncology to neurology. In addition, a critical, recent technological development is high-throughput sequencing of nucleic acids. The convergence of modern biotechnology with developments in RNA biology presents opportunities in both basic research and medical settings. Here I present two novel methods for leveraging high-throughput sequencing in the study of short non- coding RNAs, as well as a study in which they are applied to Alzheimer's Disease (AD). The computational methods presented here include High-throughput Annotation of Modified Ribonucleotides (HAMR), which enables researchers to detect post-transcriptional covalent modifications ot RNAs in a high-throughput manner. In addition, I describe Classification of RNAs by Analysis of Length (CoRAL), a computational method that allows researchers to characterize the pathways responsible for short non-coding RNA biogenesis. -
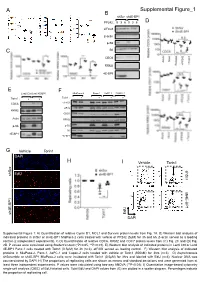
H Supplemental Figure 1 I D E B a C
A Supplemental Figure_1 B shScr sh4E-BP1 PP242 : 0 3 6 0 3 6 D eIF4G1 β-actin p-S6 C S6 CDC6 RRM2 4E-BP1 E F Lenti Ctrl Lenti 4E-BP1 MiaPaca-2 Panc-1 AsPC-1 Capan-2 Torin1 : Torin1 : + + + + + + CDC6 eIF4G1 eIF3a RRM2 CDC6 Actin RRM2 p-S6 p-S6 S6 4E-BP1 4E-BP1 G Vehicle Torin1 DAPI H I Vehicle Torin1 62.20% 45.30% shScr sh4E-BP1 EdU shScr 59.58% 49.47% EdU sh4E-BP1 DAPI Supplemental Figure 1: A) Quantification of relative Cyclin D1, MCL1 and Survivin protein levels from Fig. 1A. B) Western blot analysis of indicated proteins in shScr or sh4E-BP1 MiaPaca-2 cells treated with vehicle or PP242 (5µM) for 3h and 6h. β-actin served as a loading control (2 independent experiments). C-D) Quantification of relative CDC6, RRM2 and CDC7 protein levels from (C) Fig. 2C and (D) Fig. 2D. P values were calculated using Student’s t-test (*P<0.05, **P<0.01). E) Western blot analysis of indicated proteins in Lenti Ctrl or Lenti 4E-BP1 Panc-1 cells treated with Torin1 (0.5µM) for 3h (n=3). eIF4GI served as loading control. F) Western blot analysis of indicated proteins in MiaPaca-2, Panc-1, AsPC-1 and Capan-2 cells treated with vehicle or Torin1 (500nM) for 3hrs (n=3). G) Asynchronous shScramble or sh4E-BP1 MiaPaca-2 cells were incubated with Torin1 (0.5μM) for 3hrs and labeled with EdU (n=3). Nuclear DNA was counterstained by DAPI. H) The proportions of replicating cells are shown as means and standard deviations and were generated from at least three independent experiments. -

Molecular and Genetic Medicine
Bertazzi et al., J Mol Genet Med 2015, 8:2 Molecular and Genetic Medicine http://dx.doi.org/10.4172/1747-0862.1000116 Review Article Open Access Myotubularin MTM1 Involved in Centronuclear Myopathy and its Roles in Human and Yeast Cells Dimitri L. Bertazzi#, Johan-Owen De Craene# and Sylvie Friant* Department of Molecular and Cellular Genetics, UMR7156, Université de Strasbourg and CNRS, France #Authors contributed equally to this work. *Corresponding author: Friant S, Department of Molecular and Cellular Genetics, UMR7156, Université de Strasbourg and CNRS, 67084 Strasbourg, France, E-mail: [email protected] Received date: April 17, 2014; Accepted date: July 21, 2014; Published date: July 28, 2014 Copyright: © 2014 Bertazzi DL, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Mutations in the MTM1 gene, encoding the phosphoinositide phosphatase myotubularin, are responsible for the X-linked centronuclear myopathy (XLCNM) or X-linked myotubular myopathy (XLMTM). The MTM1 gene was first identified in 1996 and its function as a PtdIns3P and PtdIns(,5)P2 phosphatase was discovered in 2000. In recent years, very important progress has been made to set up good models to study MTM1 and the XLCNM disease such as knockout or knockin mice, the Labrador Retriever dog, the zebrafish and the yeast Saccharomyces cerevisiae. These helped to better understand the cellular function of MTM1 and of its four conserved domains: PH-GRAM (Pleckstrin Homology-Glucosyltransferase, Rab-like GTPase Activator and Myotubularin), RID (Rac1-Induced recruitment Domain), PTP/DSP (Protein Tyrosine Phosphatase/Dual-Specificity Phosphatase) and SID (SET-protein Interaction Domain). -

Metastatic Adrenocortical Carcinoma Displays Higher Mutation Rate and Tumor Heterogeneity Than Primary Tumors
ARTICLE DOI: 10.1038/s41467-018-06366-z OPEN Metastatic adrenocortical carcinoma displays higher mutation rate and tumor heterogeneity than primary tumors Sudheer Kumar Gara1, Justin Lack2, Lisa Zhang1, Emerson Harris1, Margaret Cam2 & Electron Kebebew1,3 Adrenocortical cancer (ACC) is a rare cancer with poor prognosis and high mortality due to metastatic disease. All reported genetic alterations have been in primary ACC, and it is 1234567890():,; unknown if there is molecular heterogeneity in ACC. Here, we report the genetic changes associated with metastatic ACC compared to primary ACCs and tumor heterogeneity. We performed whole-exome sequencing of 33 metastatic tumors. The overall mutation rate (per megabase) in metastatic tumors was 2.8-fold higher than primary ACC tumor samples. We found tumor heterogeneity among different metastatic sites in ACC and discovered recurrent mutations in several novel genes. We observed 37–57% overlap in genes that are mutated among different metastatic sites within the same patient. We also identified new therapeutic targets in recurrent and metastatic ACC not previously described in primary ACCs. 1 Endocrine Oncology Branch, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA. 2 Center for Cancer Research, Collaborative Bioinformatics Resource, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA. 3 Department of Surgery and Stanford Cancer Institute, Stanford University, Stanford, CA 94305, USA. Correspondence and requests for materials should be addressed to E.K. (email: [email protected]) NATURE COMMUNICATIONS | (2018) 9:4172 | DOI: 10.1038/s41467-018-06366-z | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/s41467-018-06366-z drenocortical carcinoma (ACC) is a rare malignancy with types including primary ACC from the TCGA to understand our A0.7–2 cases per million per year1,2. -

Prioritization of Metabolic Genes As Novel Therapeutic Targets in Estrogen-Receptor Negative Breast Tumors Using Multi-Omics Data and Text Mining
www.oncotarget.com Oncotarget, 2019, Vol. 10, (No. 39), pp: 3894-3909 Research Paper Prioritization of metabolic genes as novel therapeutic targets in estrogen-receptor negative breast tumors using multi-omics data and text mining Dinesh Kumar Barupal1,*, Bei Gao1,*, Jan Budczies2, Brett S. Phinney4, Bertrand Perroud4, Carsten Denkert2,3 and Oliver Fiehn1 1West Coast Metabolomics Center, University of California, Davis, CA, USA 2Institute of Pathology, Charité University Hospital, Berlin, Germany 3German Institute of Pathology, Philipps-University Marburg, Marburg, Germany 4UC Davis Genome Center, University of California, Davis, CA, USA *Co-first authors and contributed equally to this work Correspondence to: Oliver Fiehn, email: [email protected] Keywords: set-enrichment; ChemRICH; multi-omics; metabolic networks; candidate gene prioritization Received: March 12, 2019 Accepted: May 13, 2019 Published: June 11, 2019 Copyright: Barupal et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Estrogen-receptor negative (ERneg) breast cancer is an aggressive breast cancer subtype in the need for new therapeutic options. We have analyzed metabolomics, proteomics and transcriptomics data for a cohort of 276 breast tumors (MetaCancer study) and nine public transcriptomics datasets using univariate statistics, meta- analysis, Reactome pathway analysis, biochemical network mapping and text mining of metabolic genes. In the MetaCancer cohort, a total of 29% metabolites, 21% proteins and 33% transcripts were significantly different (raw p <0.05) between ERneg and ERpos breast tumors. -

Application of Microrna Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease
Article Application of microRNA Database Mining in Biomarker Discovery and Identification of Therapeutic Targets for Complex Disease Jennifer L. Major, Rushita A. Bagchi * and Julie Pires da Silva * Department of Medicine, Division of Cardiology, University of Colorado Anschutz Medical Campus, Aurora, CO 80045, USA; [email protected] * Correspondence: [email protected] (R.A.B.); [email protected] (J.P.d.S.) Supplementary Tables Methods Protoc. 2021, 4, 5. https://doi.org/10.3390/mps4010005 www.mdpi.com/journal/mps Methods Protoc. 2021, 4, 5. https://doi.org/10.3390/mps4010005 2 of 25 Table 1. List of all hsa-miRs identified by Human microRNA Disease Database (HMDD; v3.2) analysis. hsa-miRs were identified using the term “genetics” and “circulating” as input in HMDD. Targets CAD hsa-miR-1 Targets IR injury hsa-miR-423 Targets Obesity hsa-miR-499 hsa-miR-146a Circulating Obesity Genetics CAD hsa-miR-423 hsa-miR-146a Circulating CAD hsa-miR-149 hsa-miR-499 Circulating IR Injury hsa-miR-146a Circulating Obesity hsa-miR-122 Genetics Stroke Circulating CAD hsa-miR-122 Circulating Stroke hsa-miR-122 Genetics Obesity Circulating Stroke hsa-miR-26b hsa-miR-17 hsa-miR-223 Targets CAD hsa-miR-340 hsa-miR-34a hsa-miR-92a hsa-miR-126 Circulating Obesity Targets IR injury hsa-miR-21 hsa-miR-423 hsa-miR-126 hsa-miR-143 Targets Obesity hsa-miR-21 hsa-miR-223 hsa-miR-34a hsa-miR-17 Targets CAD hsa-miR-223 hsa-miR-92a hsa-miR-126 Targets IR injury hsa-miR-155 hsa-miR-21 Circulating CAD hsa-miR-126 hsa-miR-145 hsa-miR-21 Targets Obesity hsa-mir-223 hsa-mir-499 hsa-mir-574 Targets IR injury hsa-mir-21 Circulating IR injury Targets Obesity hsa-mir-21 Targets CAD hsa-mir-22 hsa-mir-133a Targets IR injury hsa-mir-155 hsa-mir-21 Circulating Stroke hsa-mir-145 hsa-mir-146b Targets Obesity hsa-mir-21 hsa-mir-29b Methods Protoc.