Pre-Processing Visualization of Hyperspectral Fluorescent
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SEVIRI HRV Fog RGB Quick Guide
SEVIRI HRV Fog RGB Quick Guide Aim: Distinguishing of fog/low clouds from snow-covered land in high resolution. Area and time period of its main application: Mid-latitude region, daytime during winter. Application and guidelines: The identification of foggy and low cloud-covered areas is important for traffic and aviation security. Winter fog/stratus frequently forms over snowy cloud-free surfaces in high pressure conditions. Fog/water clouds have good colour contrast against snow and snow-free land. This is a daytime RGB as it uses shortwave channels. SEVIRI HRV Fog RGB for 7 March 2014, 08:40 UTC Background The table shows which channels are used in the HRV Fog RGB and lists some of the land and cloud features which typically make a low or high contribution to the colour beams in this RGB. Snow's reflectivity is much higher in the HRV than in the NIR1.6. As water clouds reflect much of the radiation in both channels they can be used in combination to distinguish snow and water clouds. Smaller contribution to Larger contribution to Colour Channel [µm] Physically relates to the signal of the signal of Cloud phase Ice cloud Red NIR1.6 Water clouds Snow reflectivity Snow covered land Cloud optical thickness Thick clouds Green HRV Thin clouds Snow reflectivity Snow covered land Cloud optical thickness Thick clouds Blue HRV Thin clouds Snow refllectivity Snow covered land Notation: HRV: High Resolution Visible channel, NIR: near-infrared, number: central wavelength of the channel in micrometer. HRV is used in two colour beams so the high resolution is not lost. -

TWINSIX 2018 Sim.Pdf
@TWINSIX Due to years of customer feedback requesting the same designs between men’s and women’s jersey collections, we are offering all of the following graphics in both cuts. MEN’S & WOMEN’S ALL ITEMS MADE IN THE USA The 2018 Collection PREMIUM GEAR THE STANDARD BLACK BLUE MEN 150212-01 WOMEN 150612-01 MEN’S ONLY 170212-01 WOMEN 170612-01 WHITE OLIVE GREEN MEN 150212-02 WOMEN 150612-02 MEN’S ONLY 170212-02 WOMEN 170612-02 ORANGE KELLY GREEN MEN 150212-03 WOMEN 150612-03 MEN’S ONLY 130212-01 WOMEN 130612-01 ALL ITEMS MADE IN THE USA The 2018 Collection MEN’S & WOMEN’S GRAPHIC JERSEYS THE REVERB THE CRYPSIS WHITE, BRIGHT RED, BLACK AND TEAL BLACK WITH SHADES OF ARMY GREEN MEN 180201-10 MEN 180201-02 WOMEN 180601-10 WOMEN 180601-02 THE SURGE THE SOLOIST BLACK AND WHITE BRIGHT RED, DARK RED, BLACK, DARK BLUE AND ROYAL BLUE MEN 180201-12 MEN 180201-11 WOMEN 180601-12 WOMEN 180601-11 ALL ITEMS MADE IN THE USA The 2018 Collection MEN’S & WOMEN’S GRAPHIC JERSEYS THE POWER OF SIX THE H.C. BLACK AND OLIVE GREEN SHADES OF CYAN BLUE AND BLACK MEN 180201-09 MEN 180201-05 WOMEN 180601-09 WOMEN 180601-05 THE UPROAR THE NOMAD BLACK, WHITE AND MINT GREEN HIGH-VIZ YELLOW, RED, ROYAL BLUE AND CYAN MEN 180201-13 MEN 180201-08 WOMEN 180601-13 WOMEN 180601-08 ALL ITEMS MADE IN THE USA The 2018 Collection MEN’S & WOMEN’S GRAPHIC JERSEYS THE MARTYR THE FREEDOM MACHINE HIGH-VIZ YELLOW AND BLACK TEAL BLUE, WHITE, BRIGHT RED AND DARK RED MEN 180201-06 MEN 180201-03 WOMEN 180601-06 WOMEN 180601-03 THE G.C. -

Quick Guide Day Land Cloud
Day Land Cloud RGB Quick Guide One Product, Many Applications This RGB (also known as “Natural Color” RGB) is extremely versatile. It shows burn scars, smoke, and large fires. It clearly differentiates between liquid and ice clouds, or surface ice/snow and low Sea Ice clouds. It shows differences in land surface between marshlands and arid regions. It washes, Fire/Smoke dries, and folds your laundry…well okay, no single tool literally does it all… Ice clouds Since this product is composed of one visible and two near IR channels, it is only available during daylight hours. The good news is that these channels are common to many sensors including: VIIRS, MODIS, AVHRR, ABI, and AHI. Day Land Cloud over northeast Alaska from SNPP VIIRS at 1947 UTC, 11 Jul 2017. Note that sea ice, water clouds, ice clouds, and smoke are all evident. Day Land Cloud RGB Recipe Band / Band Diff. Physically Relates Small contribution Large Contribution Color (µm) to… indicates… indicates… Ice-phase clouds, Dry arid land, water Red 1.61 Ice and Snow snow/ice on the surface clouds, fires Little vegetation, rocky Small ice or water Green 0.86 Vegetation or bare ground particles, strong updrafts Blue 0.64 Red Visible No clouds Water clouds Impact on Operations Limitations Separating Ice and Snow Goes to Bed at Night: on the Surface from Low RGB is composed of three Clouds: low clouds composed reflectance channels of liquid water will appear requiring incoming sunshine. white while surface snow/ice will be shades of cyan. Snow and ice on the surface appear similar to cirrus clouds: Since both high level cirrus and Five-Alarm Fire: salmon color indicates large fires, surface ice/snow are frozen water they present a blue-grey streaks indicate smoke, and dark brown similar cyan color. -

Colormap Set and Get the Current Colormap
MATLAB Function Reference colormap Set and get the current colormap Syntax colormap(map) colormap(’default’) cmap = colormap Description A colormap is an m−by−3 matrix of real numbers between 0.0 and 1.0. Each row is an RGB vector that defines one color. Thek th row of the colormap defines the kth color, where map(k,:) = [r(k) g(k) b(k)]) specifies the intensity of red, green, and blue. colormap(map) sets the colormap to the matrix map. If any values in map are outside the interval [0 1], MATLAB returns the errorColormap must have values in [0,1]. colormap(’default’) sets the current colormap to the default colormap. cmap = colormap retrieves the current colormap. The values returned are in the interval [0 1]. Specifying Colormaps M−files in the color directory generate a number of colormaps. Each M−file accepts the colormap size as an argument. For example, colormap(hsv(128)) creates an hsv colormap with 128 colors. If you do not specify a size, MATLAB creates a colormap the same size as the current colormap. Supported Colormaps MATLAB supports a number of built−in colormaps, illustrated and described below. In addition to specifying built−in colormaps programmatically, you can use the Colormap menu in the Figure Properties pane of the Plot Tools GUI to select one interactively. The named built−in colormaps are the following: autumn varies smoothly from red, through orange, to yellow. bone is a grayscale colormap with a higher value for the blue component. This colormap is useful for adding an "electronic" look to grayscale images. -

Color Separation in Two-Color Printing
52 MAPS 33 Jan van de Craats Color separation in two-color printing Abstract black letters. The Graphics Companion mentions this The book Basisboek wiskunde (‘Basic Mathematics’) problem on page 348 and states that printers solve this by Jan van de Craats and Rob Bosch[1] was typeset in by trapping, which means that the size of the colored LaTEX and submitted for printing as one big pdf-file. areas is slightly increased. It doesn’t say, however, how In this book one extra color (blue) was used for titles, to achieve this in LaT X. Before explaining our simple headings, footings, important formulas, figures and E trick that does the job in a different way, let me first also as a background color for certain pages or parts of text. Jan van de Craats, who did the typesetting, devote a few words to color separation in general. reports on a trick for obtaining color separation without flaws. Color separation Color printing usually is a four step process, each page Keywords being printed four times with proportions of the ba- color, separation, printing sic colors cyan, magenta, yellow and black, respectively. Color separation from a source file consists in produ- The typescript of our Basisboek wiskunde originally was cing four distinct files in which the colors are separated prepared with a black and white text in mind. How- so that, e.g., the cyan file only contains the proportions ever, our publisher, Pearson Education Benelux, urged of cyan that should be printed. You can do color separ- us to use one extra color to enliven the general appear- ation yourself using the aurora package together with ance of the book. -

Artwork Submission Guidelines
Artwork Submission Guidelines Although it is possible to reproduce your item from a print or sample, a digital file is always preferred to help ensure that your artwork is reproduced with the greatest accuracy possible. Below are our preferred file formats for artwork submission. Please review and follow these guidelines. Preferred File Formats: VECTOR formats Vector-based PDF, EPS, AI (also DWG, DXF) • For single-ink designs or multiple spot colors • Files saved as PDF from Adobe Illustrator (best), EPS, AI • Computer-aided design & drafting (CADD) files in .DWG format (CAD files will still need extra time for setting up in house, but are better options than raster formats) Vector files represent shapes Pantone swatches are preferred for spot colors. If process colors, mathematically, providing use CMYK mode. Please create outlines of your text, as we may not perfect lines and curves. have access to the fonts (type styles) used to create the file. Vector art can be scaled to any size without affecting its Vector formats are preferred in most cases. appearance or image quality. Custom shapes: Only vector files will provide us with accurate die Vector art is preferred for lines for precise cutting. most applications, especially jobs printed with spot colors, Pre-press modification: Small adjustments are routinely made to or requiring precise cutting ensure your artwork prints as expected and conforms to the tolerances of our printing processes. Artwork which is not supplied of custom shapes. in a vector format must often be redrawn in our art department. Vector art reduces turnaround time and increases accuracy. -

Essays on Colour
Essays on Colour ESSAYS ON COLOUR A collection of columns from Cabinet Magazine Eleanor Maclure Introduction For every issue the editors of Cabinet Magazine, an American quarterly arts and culture journal, ask one of their regular contributors to write about a specific colour. The essays are printed as Cabinet’s regular Colours Column. To date, forty-two different colours have been the subject of discussion, beginning with Bice in their first ever issue. I first encountered Cabinet magazine when I stumbled upon Darren Wershler-Henry’s piece about Ruby, on the internet. I have since been able to collect all of the published columns and they have provided a wealth of knowledge, information and invaluable research about colour and colour names. Collectively, the writings represent a varied and engaging body of work, with approaches ranging from the highly factual to the deeply personal. From the birth of his niece in Matthew Klam’s Purple, to a timeline of the history of Lapis Lazuli mining in Ultramarine by Matthew Buckingham, the essays have provided fascinating insights into a whole range of colours, from basic terms such as black and red, to the more obscure: porphyry and puce. While some focus very much on the colour in question, others diverge into intricate tales of history, chemistry or geopolitics. There are personal anecdotes, legends and conspiracies, but more than that, the essays demonstrate the sheer diversity of ways we can talk about colour. The essays gathered here have become far more than just the background reading they began as. The aim of this book is to bring together the works, as a unique representation of the different ways we relate to, experience and interpret colours. -

STRATEGIC MESSAGING Brand Standards Manual 2012
STRATEGIC MESSAGING Brand Standards Manual 2012 Fernco Quality Management Systems Fernco Quality Management Systems TABLE OF CONTENTS Purpose ............................................................................................................................... 2 Fernco Brands ............................................................................................ 26 Overarching Brand/Marketing Startegy .............................................3 PlumbQwik ......................................................................................27-32 Overarching Audience Message ............................................................4 QwikSeal ..........................................................................................33-37 Brand Message .............................................................................................5 StormDrain Plus .............................................................................38-44 General Rules ............................................................................................6-7 Proflex ...............................................................................................45-50 Corporate Website/Privacy Policy .................................................. 8-11 Pow-R Repair Products ................................................................51-58 Fernco Brand .............................................................................................. 12 Wax Free Toilet Seal ......................................................................59-60 -

Download Sky Color Free Ebook
SKY COLOR DOWNLOAD FREE BOOK Peter H Reynolds | 28 pages | 01 Nov 2012 | Candlewick Press, U.S. | 9780763623456 | English | Massachusetts, United States Turquoise (color) Caribbean current. Thickening of cloud cover or the invasion of a higher cloud deck is indicative of rain in the near future. Jun 30, Tj Shay rated it it was amazing. Enlarge cover. View Sky Color comments. Light green. Fantetti e C. His illustrations are wonderful and he has a way of telling a simple story in an inspiring and artful way. The Sun and sometimes the Moon are visible in the daytime sky unless obscured by clouds. Midnight green. A typical sample is shown for each name; a range of color-variations is commonly associated with each color-name. Fantetti and C. Sky blue is a colour that resembles the colour of the unclouded sky at noon azure reflecting off a metallic surface. I would recommend this book for K Shades of cyan. Welcome back. The night sky and studies of it have a historical place in Sky Color ancient and modern cultures. Reynolds presents the story of Marisol in this third book in what the dust-jacket blurb calls his "Creatrilogy. Reynolds has created yet another gorgeous book about being yourself and finding your inner artist! Namespaces Article Talk. Skywatch: Signs of the Weather. Sinauer Associates, Inc. About Peter H. Will she figure out how to draw the sky without blue? I appreciate her activism and how she cares about things. Specifically, the colour Capri is named after the colour of the Blue Grotto on the island of Capri. -
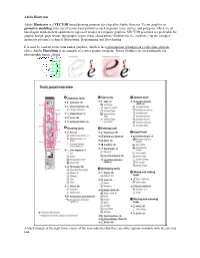
Adobe Illustrator Adobe Illustrator Is a VECTOR-Based Drawing Program Developed by Adobe Systems. Vector Graphics Or Geometric M
Adobe Illustrator Adobe Illustrator is a VECTOR-based drawing program developed by Adobe Systems. Vector graphics or geometric modeling is the use of geometrical primitives such as points, lines, curves, and polygons, which are all based upon mathematical equations to represent images in computer graphics. VECTOR programs are preferable for graphic design, page layout, typography, logos, sharp-edged artistic illustrations (i.e. cartoons, clip art, complex geometric patterns), technical illustrations, diagramming and flowcharting. It is used by contrast to the term raster graphics, which is the representation of images as a collection of pixels (dots). Adobe PhotoShop is an example of a raster graphic program. Raster Graphics are used primarily for photographic image editing. A black triangle at the right lower corner of the icon indicates that there are other options available with the selected tool. The first two tools on the toolbar are your selection tools. The black arrow will “SELECT ALL” meaning if you click on an object, the entire object will be selected including all points. The white arrow by contrast is a DIRECT SELECT tool. This tool will only select one point or if you extend a marquee, several points. The pen tool has 3 additional options. This tool is used to create accurate lines. To create straight lines, just click and release to place points. A line will be created between the points you create. Another type of line that can be created using the pen tool is the curved line. The curve that is created is known as a Bezier curve. Pronounced bez-ee-ay, these curved lines (splines) are defined by mathematical formulas. -

Displaymaker Legacy Colormark+ User Guide
DisplayMaker Legacy ColorMark+ User Guide Part Number 0706133 Rev C 1 Legal notices © Copyright 2008 Hewlett-Packard Development Company, L.P. Confidential computer software. Valid license from HP required for possession, use or copying. Consistent with FAR 12.211 and 12.212, Commercial Computer Software, Computer Software Documentation, and Technical Data for Commercial Items are licensed to the U.S. Government under vendor’s standard commercial license. The information contained herein is subject to change without notice. The only warranties for HP products and services are set forth in the express warranty statements accompanying such products and services. Nothing herein should be construed as constituting an additional warranty. HP shall not be liable for technical or editorial errors or omissions contained herein. Printed in the US For additional technical support and user documentation please refer to: www.hp.com/go/graphicarts 2 Revision Log See the accompanying Release Notes for specific changes to the software and hardware between releases of the manual. Date Description May 1999 Initial release. Aug 1999 Revised for version 1.1. Chapter 2 documents exten- sive changes to the Profile Creation Wizard, and new support for the ColorMark Calibrator. Sep 2000 Revised for version 2.0. Chapters 2 and 4 replaced with new procedures. Chapter 5 updated with new features that were added to version 1.5. Dec 2001 Revised for version 3.0. Chapter 2 adds screen dia- grams for revised dialog boxes. Chapter 3 adds information on FTP transfer of color profiles, adds details on ICC workflows, adds table of compatible applications. Chapter 4 explains the redesigned pro- file editing process. -

Efinomics: Achieving Flawless Print Quality Four Key Ingredients… and One Innovative Inkjet Printing System
EFInomics: Achieving flawless print quality Four key ingredients… And one innovative inkjet printing system EFI™ VUTEk® LX3 Pro Awesome Color Four color printing can now replace eight color modes most of the time, without sacrificing color gamut. Awesome Color And when you print with four colors, you’re using less ink and saving money! Amazing Grayscale Grayscale… it’s now your go to mode over binary, because it delivers better quality with near 1000 dpi quality at 600 dpi speeds. Amazing Grayscale It reduces unwanted visual artifacts for better color. And it’s all done with a dot LUT in our Fiery digital front end. Superior Design With the help of some pretty innovative design improvements, the visual artifacts associated with traditional inkjet printing are diminished. Superior Design Advancements in rail and bearing designs resulted in less artifacts in the printed area. Superior Design Our pinned jet plate reduces pass-to-pass banding and gloss banding. Superior Design And improved velocity alignment methods means better head-to-head drop placement for reduced graininess and banding. Genuine EFI Inks Our color scientists developed new shades of cyan, magenta and yellow pigments that, along with Fiery® XF color management tools, extend the color gamut of Genuine EFI Inks. Genuine EFI Inks With 10% to 15% more gamut, you get better Pantone® color match without giving up other ink channels to alternative ink colors. Add it all up… … and you get the new standard for image quality. Don’t let the limitations of your existing print technology be the rapid, disruptive and unpredictable changes in your business.