An Ontology of Human Developmental Anatomy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

When Does Human Life Begin? the Scientific Evidence and Terminology Revisited
University of St. Thomas Journal of Law and Public Policy Volume 8 Issue 1 Fall 2013 Article 4 January 2013 When Does Human Life Begin? The Scientific videnceE and Terminology Revisited Maureen L. Condic Follow this and additional works at: https://ir.stthomas.edu/ustjlpp Part of the Family Law Commons, and the Health Law and Policy Commons Recommended Citation Maureen L. Condic, When Does Human Life Begin? The Scientific videnceE and Terminology Revisited, 8 U. ST. THOMAS J.L. & PUB. POL'Y 44 (2013). Available at: https://ir.stthomas.edu/ustjlpp/vol8/iss1/4 This Article is brought to you for free and open access by UST Research Online and the University of St. Thomas Journal of Law and Public Policy. For more information, please contact the Editor-in-Chief at [email protected]. WHEN DOES HUMAN LIFE BEGIN? THE SCIENTIFIC EVIDENCE AND TERMINOLOGY REVISITED MAUREEN L. CONDIC* The question of when human life begins continues to be a source of ethical and political controversy. In this debate, the language used by many medical textbooks fosters significant misinterpretation of the scientific facts. In particular, terminology that refers to the product of sperm-egg fusion as a "penetrated oocyte" and claims that the zygote does not form until syngamy (approximately twenty-four hours after sperm-egg fusion) have resulted in the erroneous belief that a human embryo does not exist during the period prior to this point (i.e. the "pre-zygote error"). Yet an objective view of the modem scientific evidence supports only a single definition of the term "zygote": a one-cell human organism (i.e. -

Introduction – Developmental Overview of the Human Embryo
1 Introduction – Developmental Overview of the Human Embryo Shigehito Yamada1, and Tetsuya Takakuwa2 1Congenital Anomaly Research Center, Kyoto University, 2Human Health Science, Kyoto University, Japan 1. Introduction In this chapter, we provide a historical background on human embryo collections and describe their significant contribution to the understanding of human ontogenesis. More particularly, an overview of human embryonic development is presented using computer- generated images obtained from embryonic specimens housed at the Kyoto Collection in Japan. 1.1 Human embryology and embryo collections Historically, several human embryo collections have been created. The Carnegie Collection, the Blechschmidt Collection, the Hinrichsen Collection and the Kyoto Collection are reported as the four famous compendiums of human embryos in the world. The Carnegie Collection is the oldest and was established as early as 1887, while the Blechschmidt collection was created in 1948 by the Göttingen anatomist Erich Blechschmidt, well known for its contribution to the development of novel methods of reconstruction. In 1961, the Kyoto Collection of Human Embryos was instigated, followed by the Hinrichsen Collection in 1969. While the Blechschmidt and the Hinrichsen collections are described in Chapter 2, here, we focus on the Carnegie and the Kyoto Collections. 1.2 The carnegie human embryo collection The basis of the Carnegie Human Embryo Collection was established by Franklin P. Mall. After earning his medical degree at the University of Michigan in 1883, Mall traveled to Germany to receive a clinical training and there he met Wilhelm His and other eminent biologists. Mall then became aware of the importance of studying human embryology, and initiated a collection of human embryos in 1887. -

Human Development Domain of the Ontology of Craniofacial
Human Development Domain of the Ontology of Craniofacial Development and Malformation Jose LV Mejino Jr.1.*, Ravensara S Travillian1,2,, Timothy C Cox3,4,5., Linda G Shapiro1,2,6 and James F Brinkley1,2 1 Structural Informatics Group, Department of Biological Structure, University of Washington, Seattle, WA USA 2 Department of Biomedical Informatics and Medical Education, University of Washington, Seattle, WA USA 3 Division of Craniofacial Medicine, Department of Pediatrics, University of Washington, Seattle, WA USA 4 Center for Tissue and Cell Sciences, Seattle Children’s Research Institute; Seattle, WA USA 5Department of Anatomy & Developmental Biology, Monash University, Clayton, Victoria, Australia 6Department of Computer Science & Engineering, University of Washington, Seattle, WA USA ABSTRACT computer-processable way. The ATA component of the In this paper we describe an ontological scheme for representing ana- FMA was not built as part of the original ontology, but a tomical entities undergoing morphological transformation and changes in phenotype during prenatal development. This is a proposed component of need for it has come up in our current work on the design the Anatomical Transformation Abstraction (ATA) of the Foundational and implementation of the Ontology of Craniofacial Devel- Model of Anatomy (FMA) Ontology that was created to provide an onto- opment and Malformation (OCDM), which we are building logical framework for capturing knowledge about human development from the zygote to postnatal life. It is designed to initially describe the as part of our NIDCR-sponsored project for the FaceBase structural properties of the anatomical entities that participate in human Consortium (https://www.facebase.org/) [4], which deals development and then enhance their description with developmental prop- with craniofacial abnormalities. -

Embryo Makes Contact with the Endometrial Lining of the Uterus
Week 1 • Week 1 - Early zygote • Stage 1 starts at the beginning of • Week 1 Carnegie stage – 1,2,3,4, fertilization • Fertilization • Stage 2 begins with the division • of the zygote into two cells and Zygote ends with the appearance of the • Morula blastocystic cavity • Blastocyst • Stage 3 begins when the blastocystic cavity first appears in the morula and ends when the zona (capsula) pellucida is shed as the embryo makes contact with the endometrial lining of the uterus. • Stage 4 is reserved for the attaching blastocyst to the endometrial lining Week 2 • Week 2 Implantation • Stage 5 Two distinct layers • Week 2 Carnegie stage -5,6 are evident in the • Trophoblast - outer cell trophoblast; 1) a thicker layer outer layer without cell boundaries, called the • Embryoblast - inner cell syncytiotrophoblast and 2) mass a thinner inner layer with • Implantation cell boundaries called the • Bilaminar embryo cytotrophoblast. • Stage 6 the first appearance of chorionic villi. Week 3 • • Stage 7 the presomite • Week 3 - Embryonic disc period and well defined • Week 3 - Carnegie stage – embryonic disc appearance 7,8, &9 of the notochordal process and the gastrulation • Gastrulation (primitive) node. • Notochord formation • Trilaminar embryo • Mesoderm • Somitogenesis • Neurogenesis Week 4 • The heart begins • Week 4 - Carnegie stage -10,11,12 &13 • Heart • Placodes • Pharyngeal arches • Week 5 • Week 7 - Head and limb • Carnegie stages stage 14 development stage 15 • Carnegie stages stage 18 • stage 19 • Week 6 - Early face • Week 8 deevelopment • Last embryonic stage • Carnegie stages Carnegie stage – 20 21 22 • Week 6 - Carnegie stage 16 &23 & 17 • Last week of embryonic development. -

Embryology and Fetal Development 2016.Pages
page 1 Embryology and Fetal Development Study Group Module Learning Objectives Review the following Learning Objectives as an organized beginning to your study of this module. As you read the Learning Objectives, note key words that will aid you in finding the information in the texts. When you complete the module, revisit this list and check for areas that require further investigation. • Review the mechanisms of conception. • Identify the difference between gestational age and dating from LMP. • Identify the Carnegie Stages of Development • Discuss the real-life considerations regarding habit, nutrition, and lifestyle that can impact a developing embryo. • Identify the rapidly changing occurrences in cell division and development from conception to the beginning of the embryonic period. • Identify the embryonic and fetal periods. • Identify the process of twinning. • Identify the changes seen in early cell specialization through the formation of the ectoderm, mesoderm and endoderm germ layers. • Identify the early formation and development of the neural tube, chorionic villi, endocrine and renal systems. • Understand embryonic/fetal circulation and development. • Understand embryonic/fetal liver development. • Identify the growth and development of the embryonic skeletal structure. • Identify the major changes during the fetal development period, including major organ systems growth and development. • Identify the role of X and Y-chromosomes and the SRY gene. • Identify the approximate size of the developing fetus during the passing weeks of gestation. • Review the maturation of fetal lungs. • Identify the fetal brain development that occurs between 35 and 40 weeks. • Understand the timing of exposure to teratogens and the potential effects on a developing embryo and fetus. -

Findings from Chick Embryo Studies
Journal of Cardiovascular Development and Disease Review Mechanosensitive Pathways in Heart Development: Findings from Chick Embryo Studies Maha Alser 1,2, Samar Shurbaji 1 and Huseyin C. Yalcin 1,* 1 Biomedical Research Center, Qatar University, Doha P.O. Box 2713, Qatar; [email protected] (M.A.); [email protected] (S.S.) 2 College of Health and Life Sciences, Hamad Bin Khalifa University, Doha P.O. Box 34110, Qatar * Correspondence: [email protected]; Tel.: +974-4403-7719 Abstract: The heart is the first organ that starts to function in a developing embryo. It continues to undergo dramatic morphological changes while pumping blood to the rest of the body. Genetic regulation of heart development is partly governed by hemodynamics. Chick embryo is a major animal model that has been used extensively in cardiogenesis research. To reveal mechanosensitive pathways, a variety of surgical interferences and chemical treatments can be applied to the chick embryo to manipulate the blood flow. Such manipulations alter expressions of mechanosensitive genes which may anticipate induction of morphological changes in the developing heart. This paper aims to present different approaches for generating clinically relevant disturbed hemodynamics conditions using this embryonic chick model and to summarize identified mechanosensitive genes using the model, providing insights into embryonic origins of congenital heart defects. Keywords: chick embryo; hemodynamics; mechanobiology; mechanosensitive pathways; surgical interferences; left atrial ligation; outflow tract banding; vitelline vein ligation; chemical treatment; gene expression Citation: Alser, M.; Shurbaji, S.; Yalcin, H.C. Mechanosensitive Pathways in Heart Development: Findings from Chick Embryo Studies. 1. Introduction J. Cardiovasc. Dev. -

Cultured Human Pre-Gastrulation Embryos
Protocol for a developmental landscape of 3D- cultured human pre-gastrulation embryos Lifeng Xiang Yunnan Key Laboratory of Primate Biomedical Research; Institute of Primate Translational Medicine, Kunming University of Science and Technology Yu Yin Yunnan Key Laboratory of Primate Biomedical Research; Institute of Primate Translational Medicine, Kunming University of Science and Technology Tianqing Li ( [email protected] ) Yunnan Key Laboratory of Primate Biomedical Research; Institute of Primate Translational Medicine, Kunming University of Science and Technology Method Article Keywords: Human pre-gastrulation embryo, three-dimensional (3D) culture, primitive streak anlage, immunouorescence imaging, single cell RNA-seq Posted Date: December 12th, 2019 DOI: https://doi.org/10.21203/rs.2.16169/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/9 Abstract Human embryogenesis is not well understood. Knowledge detailing human pre-gastrulation embryonic development including spatial self-organization and cell type ontogeny remains limited by available two- dimensional technological platforms. Here, we present a three-dimensional (3D) blastocyst-culture system, which enables human blastocyst development through primitive streak anlage (PSA). By the 3D- platform combined with immunouorescence imaging and single-cell RNA-Seq, we reveal a developmental landscape of human pre-gastrulation embryos. Our protocol allows recording and analysis of embryo developmental landmarks and mechanisms from human blastocysts to pre- gastrulation stage (day 14 post- fertilization). Introduction Early human embryogenesis, such as architecture formation and cell type specication, is obscure owing to technical challenge and unavailable materials. Recent in vitro implantation platforms using a two- dimensional (2D) culture approach have revealed some developmental landmarks of in vivo early human embryos1,2. -

Lessons from Skeletal Muscle Development
REPRODUCTIONREVIEW Many routes to the same destination: lessons from skeletal muscle development Gi Fay Mok and Dylan Sweetman Division of Animal Sciences, School of Biosciences, University of Nottingham, Sutton Bonington Campus, Loughborough, Leicestershire LE12 5RD, UK Correspondence should be addressed to D Sweetman; Email: [email protected] Abstract The development and differentiation of vertebrate skeletal muscle provide an important paradigm to understand the inductive signals and molecular events controlling differentiation of specific cell types. Recent findings show that a core transcriptional network, initiated by the myogenic regulatory factors (MRFs; MYF5, MYOD, myogenin and MRF4), is activated by separate populations of cells in embryos in response to various signalling pathways. This review will highlight how cells from multiple distinct starting points can converge on a common set of regulators to generate skeletal muscle. Reproduction (2011) 141 301–312 Introduction in origin. The locations of these areas in the embryo are shown schematically in Fig. 1A. Most muscles are derived Understanding the process of differentiation is a central from paraxial trunk mesoderm (i.e. the mesoderm lying concern of developmental biology, and the study of immediately adjacent to the midline, see Fig. 1A and B), vertebrate skeletal muscle provides an excellent para- which forms somites, and head mesoderm. Additionally, digm to understand this problem. Investigations over some lateral neck mesoderm has also been shown to have many years have shed light on the inductive processes myogenic potential. Trunk, head, neck and limb muscles within embryos that lead to myogenesis and the are all produced by distinct developmental pathways, molecular events that underpin this. -

Development, Differentiation and Derivatives of the Wolffian and Müllerian Ducts
9 Development, Differentiation and Derivatives of the Wolffian and Müllerian Ducts Monika Jacob, Faisal Yusuf and Heinz Jürgen Jacob Department of Anatomy and Molecular Biology, Ruhr-University Bochum, Germany 1. Introduction The Wolffian ducts (pro- and mesonephric ducts) are the most important and earliest structures formed during the development of the urogenital system in vertebrates including humans. The Wolffian ducts originate in the prospective cervical region of the young embryo but later migrate caudally inducing the development of the pronephric and mesonephric tubules along their migratory route. In addition to being the inducers of the first two generations of the kidney, namely the pronephros and mesonephros, the Wolffian ducts also give rise to the ureteric buds which drive the growth and differentiation of the permanent kidneys, the metanephroi. The paired ureteric bud arises as outpouching from the caudal end of the Wolffian duct and induces the epithelialisation of the metanephric blastema leading to the formation of the renal corpuscles and tubular part of the nascent metanephric kidney, while the entire collecting system consisting of the ureter, the renal pelvis, the calyces and the collecting ducts take their origin from the ureteric bud. Gender specific contributions of the Wolffian ducts amount to the induction and development of the Müllerian (paramesonephric) ducts, the anlagen of the female genital ducts, while in males, the Wolffian ducts elongate to form the epididymal ducts and the vasa deferentia. The seminal vesicles are formed during regression and transformation of the mesonephroi. The developmental significance of the Wolffian duct for the development of the excretory and genital system can be drawn from the extirpation experiments in vertebrate embryos where the absence of Wolffian ducts showed that neither kidneys, nor male or female genital ducts develop. -

Early Human Development and the Chief Sources of Information on Staged Human Embryos
EUROP. J. OBSTET. GYNEC. REPROD. BIOL., 1979,9/4,273-280 0 Elsevier/North-Holland Biomedical Press REVIEW ARTICLE Early human development and the chief sources of information on staged human embryos R. O’Rahilly Carnegie Laboratories of Embryology and Departments of Neurology and Human Anatomy, Universityof California,Davis, Calif: 95616, U.S.A. Accepted for publication 25 January 1979 O’RAHILLY, R. (1979): Early human development and the chief sources of information on staged human embryos. Europ. J. Obster. gynec. reprod. Biol., 914, 213-280. In a brief historical survey, the importance of Wilhelm His, senior, to human embryology is emphasized. He provided the impetus to Mail to establish the Carnegie Embryological Collection, which serves as a ‘Bureau of Standards’ for early human development. The Carnegie system of 23 stages for the embryonic period proper (first 8 postovulatory wk) is outlined, and some common misusages are noted. Finally, because of the difficulty in tracking down data based on staged human embryos, an annotated list of more than 40 key references is provided. embryonic period; embryonic stages; developmental ‘horizons’; Carnegie Collection; Wilhelm His, senior “The norm should be established; “provided the incentive for Keibel’s Norrnentafeh . embryos should be arranged in stages” and [for my (the future Carnegie) collection] of hu- (Franklin P. Mall) man embryos” (Mall, 1913). An important landmark, and still the most detailed account available, was the “Manual of Human Em- introduction and historical aspects bryology” -

The Neural Crest and Craniofacial Malformations
Chapter 5 The Neural Crest and Craniofacial Malformations Hans J.ten Donkelaar and Christl Vermeij-Keers 5.1 Introduction the head (Vermeij-Keers 1990; Sulik 1996; LaBonne and Bronner-Fraser 1999; Le Douarin and Kalcheim The neural crest is a temporary embryonic structure 1999; Sperber 2002; Knecht and Bronner-Fraser 2002; that is composed of a population of multipotent cells Francis-West et al. 2003; Santagati and Rijli 2003). In that delaminate from the ectoderm by epitheliomes- addition, during later developmental stages multiple enchymal tranformation (EMT; Duband et al. 1995; places of EMT are recognized as well. In the head– Hay 1995; Le Douarin and Kalcheim 1999; Francis- neck area, for example, the neurogenic placodes and West et al. 2003). Neural-crest-derived cells are called the optic neural crest are such areas. Neurogenic pla- mesectodermal or ectomesenchymal cells (mesoder- codes, specialized regions of the embryonic ecto- mal cells of ectodermal origin) that have arisen derm, are the major source of primary sensory neu- through EMT. The neural crest was first described rons in the head (Johnston and Bronsky 1995; Gra- by His (1868) in the chick embryo as a Zwischen- ham and Begbie 2000). The vasculature of the head is strang, a strip of cells lying between the dorsal ecto- derived from mesoderm-derived endothelial precur- derm and the neural tube. Classic contributions in sors,while the neural crest provides the pericytes and amphibians identified interactions between tissues smooth muscle cells of the vessels of the face and the that lead to neural crest formation, and were re- forebrain (Etchevers et al. -
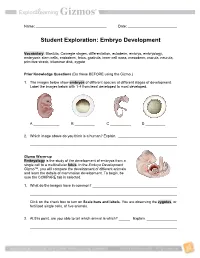
Embryo Development
Name: Date: Student Exploration: Embryo Development Vocabulary: Blastula, Carnegie stages, differentiation, ectoderm, embryo, embryology, embryonic stem cells, endoderm, fetus, gastrula, inner cell mass, mesoderm, morula, neurula, primitive streak, trilaminar disk, zygote Prior Knowledge Questions (Do these BEFORE using the Gizmo.) 1. The images below show embryos of different species at different stages of development. Label the images below with 1-4 from least developed to most developed. A B C D 2. Which image above do you think is a human? Explain. Gizmo Warm-up Embryology is the study of the development of embryos from a single cell to a multicellular fetus. In the Embryo Development Gizmo™, you will compare the development of different animals and learn the details of mammalian development. To begin, be sure the COMPARE tab is selected. 1. What do the images have in common? Click on the check box to turn on Scale bars and labels. You are observing the zygotes, or fertilized single cells, of five animals. 2. At this point, are you able to tell which animal is which? Explain. Get the Gizmo ready: Activity A: Make sure the COMPARE tab is selected and the Comparative Developmental stage slider is on stage 1. embryology Turn off Scale bars and labels. Introduction: Comparative embryology compares embryos of different species to gain insight into how they are related. To provide a framework for this comparison, embryologists divide this process into 23 Carnegie stages. You will look at seven of these stages in this activity. Question: Can we use comparative embryology to determine the relatedness of species? 1.