Stabilizing Selection Maintains Exuberant Colour Polymorphism in the Spider Theridion Californicum (Araneae, Theridiidae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Introgression Analysis of Quantitative Trait Loci That Contribute to a Morphological Difference Between Drosophila Simulans and D
Copyight 0 1997 by the Genetics Societv of America An Introgression Analysis of Quantitative Trait Loci That Contribute to a Morphological Difference Between Drosophila simulans and D. mauritiana Cathy C. Laurie, John R. True, Jianjun Liu and John M. Mercer Department of Zoology, Duke University, Durham, North Carolina 27708 Manuscript received August 12, 1996 Accepted for publication October 29, 1996 ABSTRACT Drosophila simulans and D. maum'tiana differ markedly in morphology of the posterior lobe, a male- specific genitalic structure. Bothsize and shapeof the lobe can be quantifiedby a morphometric variable, PC1, derived from principal components and Fourier analyses. The genetic architectureof the species difference in PC1 was investigated previously by composite interval mapping, which revealed largely additive inheritance, with a minimum of eight quantitative trait loci (QTL) affecting the trait. This analysis was extended by introgression of marked segments of the maun'tiana third chromosome into a simulans background by repeated backcrossing. Thetwo types of experiment are consistentin suggesting that several QTL on the third chromosome may have effects in the range of 10-15% of the parental difference andthat all or nearly all QTL have effects in the same direction. Since the parental difference is large (30.4 environmental standard deviations), effects of this magnitude can produce alternative homozygotes with little overlap in phenotype. However, these estimates may not reflect the effects of individual loci, since each interval or introgressed segment may contain multiple QTL. The consistent direction of allelic effects suggests a history of directional selection on the posterior lobe. HE genetic analysis of a quantitative trait usually ual genes that affect continuously variable traits. -

Molecular Insights Into the Phylogenetic Structure of the Spider
MolecularBlackwell Publishing Ltd insights into the phylogenetic structure of the spider genus Theridion (Araneae, Theridiidae) and the origin of the Hawaiian Theridion-like fauna MIQUEL A. ARNEDO, INGI AGNARSSON & ROSEMARY G. GILLESPIE Accepted: 9 March 2007 Arnedo, M. A., Agnarsson, I. & Gillespie, R. G. (2007). Molecular insights into the phylo- doi:10.1111/j.1463-6409.2007.00280.x genetic structure of the spider genus Theridion (Araneae, Theridiidae) and the origin of the Hawaiian Theridion-like fauna. — Zoologica Scripta, 36, 337–352. The Hawaiian happy face spider (Theridion grallator Simon, 1900), named for a remarkable abdominal colour pattern resembling a smiling face, has served as a model organism for under- standing the generation of genetic diversity. Theridion grallator is one of 11 endemic Hawaiian species of the genus reported to date. Asserting the origin of island endemics informs on the evolutionary context of diversification, and how diversity has arisen on the islands. Studies on the genus Theridion in Hawaii, as elsewhere, have long been hampered by its large size (> 600 species) and poor definition. Here we report results of phylogenetic analyses based on DNA sequences of five genes conducted on five diverse species of Hawaiian Theridion, along with the most intensive sampling of Theridiinae analysed to date. Results indicate that the Hawai- ian Islands were colonised by two independent Theridiinae lineages, one of which originated in the Americas. Both lineages have undergone local diversification in the archipelago and have convergently evolved similar bizarre morphs. Our findings confirm para- or polyphyletic status of the largest Theridiinae genera: Theridion, Achaearanea and Chrysso. -

Microevolution and the Genetics of Populations Microevolution Refers to Varieties Within a Given Type
Chapter 8: Evolution Lesson 8.3: Microevolution and the Genetics of Populations Microevolution refers to varieties within a given type. Change happens within a group, but the descendant is clearly of the same type as the ancestor. This might better be called variation, or adaptation, but the changes are "horizontal" in effect, not "vertical." Such changes might be accomplished by "natural selection," in which a trait within the present variety is selected as the best for a given set of conditions, or accomplished by "artificial selection," such as when dog breeders produce a new breed of dog. Lesson Objectives ● Distinguish what is microevolution and how it affects changes in populations. ● Define gene pool, and explain how to calculate allele frequencies. ● State the Hardy-Weinberg theorem ● Identify the five forces of evolution. Vocabulary ● adaptive radiation ● gene pool ● migration ● allele frequency ● genetic drift ● mutation ● artificial selection ● Hardy-Weinberg theorem ● natural selection ● directional selection ● macroevolution ● population genetics ● disruptive selection ● microevolution ● stabilizing selection ● gene flow Introduction Darwin knew that heritable variations are needed for evolution to occur. However, he knew nothing about Mendel’s laws of genetics. Mendel’s laws were rediscovered in the early 1900s. Only then could scientists fully understand the process of evolution. Microevolution is how individual traits within a population change over time. In order for a population to change, some things must be assumed to be true. In other words, there must be some sort of process happening that causes microevolution. The five ways alleles within a population change over time are natural selection, migration (gene flow), mating, mutations, or genetic drift. -

Patterns and Power of Phenotypic Selection in Nature
Articles Patterns and Power of Phenotypic Selection in Nature JOEL G. KINGSOLVER AND DAVID W. PFENNIG Phenotypic selection occurs when individuals with certain characteristics produce more surviving offspring than individuals with other characteristics. Although selection is regarded as the chief engine of evolutionary change, scientists have only recently begun to measure its action in the wild. These studies raise numerous questions: How strong is selection, and do different types of traits experience different patterns of selection? Is selection on traits that affect mating success as strong as selection on traits that affect survival? Does selection tend to favor larger body size, and, if so, what are its consequences? We explore these questions and discuss the pitfalls and future prospects of measuring selection in natural populations. Keywords: adaptive landscape, Cope’s rule, natural selection, rapid evolution, sexual selection henotypic selection occurs when individuals with selection on traits that affect survival stronger than on those Pdifferent characteristics (i.e., different phenotypes) that affect only mating success? In this article, we explore these differ in their survival, fecundity, or mating success. The idea and other questions about the patterns and power of phe- of phenotypic selection traces back to Darwin and Wallace notypic selection in nature. (1858), and selection is widely accepted as the primary cause of adaptive evolution within natural populations.Yet Darwin What is selection, and how does it work? never attempted to measure selection in nature, and in the Selection is the nonrandom differential survival or repro- century following the publication of On the Origin of Species duction of phenotypically different individuals. -

Gillespie, RG 1989. Diet-Induced Color Change in the Hawaiian Happy
Gillespie, R. G . 1989 . Diet-induced color change in the Hawaiian happy-face spider Theridion grallator (Araneae, Theridiidae) . J . Arachnol ., 17:171-177 . DIET-INDUCED COLOR CHANGE IN THE HAWAIIAN HAPPY-FACE SPIDER THERIDION GRALLATOR, (ARANEAE, THERIDIIDAE) Rosemary G. Gillespie' Department of Entomology, University of Hawaii, Honolulu, Hawaii 96822' USA ABSTRACT The Hawaiian happy-face spider Theridion grallator,Simon is a smalll spider, endemic to Hawaii, where it is found under leaves in the wet and mesic forests . The abdomen is pale, translucent yellow, but variable amounts of red, black or white pigment may be superimposed on this to generate a host of patterned morphs . The translucence of the abdomen may enhance crypsis against predators searching the underside of leaves ; the variability in the superimposed pattern may serve to counteract the development of a search image by the predator . The present study documents plasticity in base coloration, which can changee rapidly and markedly following ingestion of certain types of prey . This may be merely a consequence of abdominal translucence . But it is interesting to note that it adds a whole new dimension to the color polymorphism of the species . INTRODUCTION Color change in animals is a widespread phenomenon . It is generally associated with a change in physiological state . This in turn may be induced by ontogenetic or environmental changes, or stress . In spiders, color change is known to occur under a variety of circumstances . Ontogenetic modifictions are widespread, with the adult coloration being attained in the final molt (Bonnet 1933; Homann 1946; Millot 1949). More rapid changes in color pattern have been noted in spiders of the family Araneidae, which accumulate guanine beneath the cuticle during periods of starvation, thereby developing a pattern of opaque white blotches over the abdomen (Foelix 1979) . -

Formatting Your Paper for Submission in the Moorea Class Journal
UC Berkeley Student Research Papers, Fall 2012 Title Coloration in relation to ecology in the Asian spiny-backed spider, Thelacantha brevispina (Araneae, Araneidae) on Moorea, French Polynesia Permalink https://escholarship.org/uc/item/5524p5x6 Author Truong, Hua Publication Date 2012-12-12 Undergraduate eScholarship.org Powered by the California Digital Library University of California COLORATION IN RELATION TO ECOLOGY IN THE ASIAN SPINY-BACKED SPIDER, THELACANTHA BREVISPINA (ARANEAE, ARANEIDAE) ON MOOREA, FRENCH POLYNESIA HUA S. TRUONG Integrative Biology, University of California, Berkeley, California 94720 USA Abstract. Color polymorphism is a widespread phenomenon in many arthropods, including spiders. The persistence of these visible, readily measured polymorphisms over time suggests that they carry adaptive functions and thus are maintained by natural selection. Studying polymorphism is useful for examining evolution in action and the different modes of selection operating to maintain different morph patterns. The color polymorphic spiny-backed spider, Thelacantha brevispina, was surveyed on Moorea, French Polynesia to understand distribution, prey abundance, and density across coastal and forest habitats. In addition, split-brood and laboratory reciprocal transplant experiments were conducted to determine whether coloration was due to genes or a result of phenotypic plasticity. Quantitative field surveys revealed a negative relationship between spider density and prey abundance, with lower prey counts in coastal habitats but prey yielded higher energy value. Spiders were found on a variety of habitats with diverse prey species. The split-brood design showed that a genetic origin of coloration is likely with strong maternal effects and distinct color differences among sites. The reciprocal transplant confirmed the lack of a plastic response to rearing habitats. -
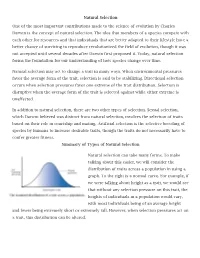
Natural Selection One of the Most Important Contributions Made to The
Natural Selection One of the most important contributions made to the science of evolution by Charles Darwin is the concept of natural selection. The idea that members of a species compete with each other for resources and that individuals that are better adapted to their lifestyle have a better chance of surviving to reproduce revolutionized the field of evolution, though it was not accepted until several decades after Darwin first proposed it. Today, natural selection forms the foundation for our understanding of how species change over time. Natural selection may act to change a trait in many ways. When environmental pressures favor the average form of the trait, selection is said to be stabilizing. Directional selection occurs when selection pressures favor one extreme of the trait distribution. Selection is disruptive when the average form of the trait is selected against while either extreme is unaffected. In addition to natural selection, there are two other types of selection. Sexual selection, which Darwin believed was distinct from natural selection, involves the selection of traits based on their role in courtship and mating. Artificial selection is the selective breeding of species by humans to increase desirable traits, though the traits do not necessarily have to confer greater fitness. Summary of Types of Natural Selection Natural selection can take many forms. To make talking about this easier, we will consider the distribution of traits across a population in using a graph. To the right is a normal curve. For example, if we were talking about height as a trait, we would see that without any selection pressure on this trait, the heights of individuals in a population would vary, with most individuals being of an average height and fewer being extremely short or extremely tall. -

Supplementary Information for Vyssotski, 2013
Supplementary Information for Vyssotski, 2013 www.evolocus.com/evolocus/v1/evolocus-01-013-s.pdf Evolocus Supplementary Information for Transgenerational epigenetic compensation and sex u al d imorph ism 1,2,3 Dmitri L. Vyssotski Published 17 May 2013, Evolocus 1, 13-18 (2013): http://www.evolocus.com/evolocus/v1/evolocus-01-013.pdf In natu ral popu lation transgenerational epigenetic compensation is by unknown mechanisms, the mechanisms those were difficult to generated in h omoz y gou s mu tants, mainly in males. It d oes not imagine, and this possibility was abandoned for over 47 years. immed iately affect th e ph enoty pe of h omoz y gou s mu tants, w h ere The higher variability in males can be a result of a not so good it w as generated . Transgenerational epigenetic compensation is canalization of their ontogenesis, and the canalization of localiz ed not in th e same locu s as original mu tation and it is ontogenesis is modulated by sex hormones. Thus, we can d ominant, b ecau se it affects th e ph enoty pe ev en if it is receiv ed imagine that the canalization of ontogenesis in males is from one parent. In fu rth er generations, starting from F 2, it is more artificially decreased in comparison with females by means of ex pressed in females th an in males: it increases fitness of hormonal differences. h omozy gou s mu tant females, d ecreases fitness of w ild ty pe The shift of the female distribution to the right was not females and h as w eak effect on h eterozy gou s females. -

Stabilizing Selection Shapes Variation in Phenotypic Plasticity
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.29.454146; this version posted July 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Stabilizing selection shapes variation in phenotypic plasticity 2 3 4 5 Dörthe Becker1,2,*, 6 Karen Barnard-Kubow1,3, 7 Robert Porter1, 8 Austin Edwards1,4, 9 Erin Voss1,5, 10 Andrew P. Beckerman2, 11 Alan O. Bergland1,* 12 13 14 1Department of Biology; University of Virginia, Charlottesville/VA, USA 15 2Department of Animal and Plant Sciences; University of Sheffield, Sheffield, UK 16 3Department of Biology, James Madison University, Harrisonburg/VA, USA 17 4Biological Imaging Development CoLab, University of California San Francisco, 18 San Francisco/CA, USA 19 5Department of Integrative Biology, University of California Berkeley, 20 Berkeley/CA 21 22 * to whom correspondence should be addressed: 23 DB ([email protected]), AOB ([email protected]) 24 25 26 27 Keywords: stabilizing selection; phenotypic plasticity; heritability; Daphnia; 28 defense morphologies 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.29.454146; this version posted July 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 29 Abstract 30 The adaptive nature of phenotypic plasticity is widely documented in natural 31 populations. However, little is known about the evolutionary forces that shape 32 genetic variation in plasticity within populations. Here we empirically address this 33 issue by testing the hypothesis that stabilizing selection shapes genetic variation 34 in the anti-predator developmental plasticity of Daphnia pulex. -

Selection on Breeding Date and Body Size in Colonizing Coho Salmon, Oncorhynchus Kisutch
Molecular Ecology (2010) 19, 2562–2573 doi: 10.1111/j.1365-294X.2010.04652.x Selection on breeding date and body size in colonizing coho salmon, Oncorhynchus kisutch J. H. ANDERSON,* P. L. FAULDS,† W. I. ATLAS,‡ G. R. PESS§ and T. P. QUINN* *School of Aquatic and Fishery Sciences, University of Washington, Box 355020, Seattle, WA 98195, USA, †Seattle Public Utilities, 700 5th Avenue, Suite 4900, Seattle, WA, 98124, USA, ‡Department of Biological Sciences, Simon Fraser University, 8888 University Drive, Burnaby, BC, Canada V5A 1S6, §National Marine Fisheries Service, Northwest Fisheries Science Center, 2725 Montlake Boulevard East, Seattle, WA 98195, USA Abstract Selection during the colonization of new habitat is critical to the process of local adaptation, but has rarely been studied. We measured the form, direction, and strength of selection on body size and date of arrival to the breeding grounds over the first three cohorts (2003–2005) of a coho salmon (Oncorhynchus kisutch) population colonizing 33 km of habitat made accessible by modification of Landsburg Diversion Dam, on the Cedar River, Washington, USA. Salmon were sampled as they bypassed the dam, parentage was assigned based on genotypes from 10 microsatellite loci, and standardized selection gradients were calculated using the number of returning adult offspring as the fitness metric. Larger fish in both sexes produced more adult offspring, and the magnitude of the effect increased in subsequent years for males, suggesting that low densities attenuated traditional size-biased intrasexual competition. For both sexes, directional selection favoured early breeders in 2003, but stabilizing selection on breeding date was observed in 2004 and 2005. -

Environmental Review 500 MW Geothermal Development
Environmental Review 500 MW Geothermal Development PRELIMINARY DRAFT Technical Description and Environmental Setting December 12, 1988 TABLE OF CONTENTS I INTRODUCTION A. OVERVIEW I-1 B. GEOTHERMAL RESOURCE POTENTIAL I-1 C. PUBLIC INVOLVEMENT I-2 D. ENVIRONMENTAL CONSIDERATIONS I-4 II. DESCRIPTION OF THE DEVELOPMENT CONCEPT A. OVERVIEW OF THE GEOTHERMAL ENERGY TO ELECTRICITY CONVERSION PROCESS II-1 B. BASIC ASSUMPTIONS II-2 C. DEVELOPMENT CONCEPT II-3 D. TECHNICAL DESCRIPTION OF THE DESIGN, CONSTRUCTION AND OPERATION OF GEOTHERMAL WELLS II-5 E. TECHNICAL DESCRIPTION OF THE DESIGN, CONSTRUCTION AND OPERATION OF GEOTHERMAL POWER PLANTS II-14 F. POWER TRANSMISSION II-24 G. CONVERTER STATION II-27 H. INFRASTRUCTURE AND UTILITIES II-27 III. ENVIRONMENTAL SETTING A. GEOLOGY AND SOILS III-a1 B. METEOROLOGY, AIR QUALITY AND NOISE III-b1 C. HYDROLOGY AND WATER QUALITY III-c1 D. CULTURAL RESOURCES III-d1 E. FLORA AND FAUNA III-e1 IV. ECONOMICS AND SOCIO-ECONOMIC SETTING A. ASSUMPTIONS IV-1 B. ECONOMIC CHARACTERISTICS IV-2 C. SOCIO-ECONOMIC SETTING IV-6 PART I: INTRODUCTION A. OVERVIEW Hawaii presently relies upon petroleum fuel to supply 90 percent of its total energy needs, making the State vulnerable to sudden shortages in supply or escalations in the price of this diminishing source of energy. A major goal for the state in the State Energy Functional Plan (OPED, 1984), is to reduce Hawaii's dependency on oil through the use of alternate forms of energy. As stated in the Functional Plan, it is a priority objective for the State to "Accelerate the transition to an indigenous renewable energy economy by facilitating private sector activities to explore supply options and achieve local commercialization and application of appropriate energy technologies." Geothermal heat as an alternative energy source was first explored for commercial use in Hawaii in 1961, when four test holes were drilled in the Kilauea East Rift Zone by a private company. -

UC Riverside UC Riverside Previously Published Works
UC Riverside UC Riverside Previously Published Works Title Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit. Permalink https://escholarship.org/uc/item/95h4x800 Journal BMC genomics, 15(1) ISSN 1471-2164 Authors Clarke, Thomas H Garb, Jessica E Hayashi, Cheryl Y et al. Publication Date 2014-05-23 DOI 10.1186/1471-2164-15-365 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Clarke et al. BMC Genomics 2014, 15:365 http://www.biomedcentral.com/1471-2164/15/365 RESEARCH ARTICLE Open Access Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit Thomas H Clarke1, Jessica E Garb2, Cheryl Y Hayashi3, Robert A Haney2, Alexander K Lancaster4,5, Susan Corbett2 and Nadia A Ayoub1* Abstract Background: Spiders (Order Araneae) are essential predators in every terrestrial ecosystem largely because they have evolved potent arsenals of silk and venom. Spider silks are high performance materials made almost entirely of proteins, and thus represent an ideal system for investigating genome level evolution of novel protein functions. However, genomic level resources remain limited for spiders. Results: We de novo assembled a transcriptome for the Western black widow (Latrodectus hesperus) from deeply sequenced cDNAs of three tissue types. Our multi-tissue assembly contained ~100,000 unique transcripts, of which > 27,000 were annotated by homology. Comparing transcript abundance among the different tissues, we identified 647 silk gland-specific transcripts, including the few known silk fiber components (e.g.