Transcriptional Changes of Salicylic Acid Dependent Signaling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Emergence of Cereal Fungal Diseases and the Incidence of Leaf Spot Diseases in Finland
AGRICULTURAL AND FOOD SCIENCE AGRICULTURAL AND FOOD SCIENCE Vol. 20 (2011): 62–73. Vol. 20(2011): 62–73. The emergence of cereal fungal diseases and the incidence of leaf spot diseases in Finland Marja Jalli, Pauliina Laitinen and Satu Latvala MTT Agrifood Research Finland, Plant Production Research, FI-31600 Jokioinen, Finland, email: [email protected] Fungal plant pathogens causing cereal diseases in Finland have been studied by a literature survey, and a field survey of cereal leaf spot diseases conducted in 2009. Fifty-seven cereal fungal diseases have been identified in Finland. The first available references on different cereal fungal pathogens were published in 1868 and the most recent reports are on the emergence of Ramularia collo-cygni and Fusarium langsethiae in 2001. The incidence of cereal leaf spot diseases has increased during the last 40 years. Based on the field survey done in 2009 in Finland, Pyrenophora teres was present in 86%, Cochliobolus sativus in 90% and Rhynchosporium secalis in 52% of the investigated barley fields.Mycosphaerella graminicola was identi- fied for the first time in Finnish spring wheat fields, being present in 6% of the studied fields.Stagonospora nodorum was present in 98% and Pyrenophora tritici-repentis in 94% of spring wheat fields. Oat fields had the fewest fungal diseases. Pyrenophora chaetomioides was present in 63% and Cochliobolus sativus in 25% of the oat fields studied. Key-words: Plant disease, leaf spot disease, emergence, cereal, barley, wheat, oat Introduction nbrock and McDonald 2009). Changes in cropping systems and in climate are likely to maintain the plant-pathogen interactions (Gregory et al. -

The Phylogeny of Plant and Animal Pathogens in the Ascomycota
Physiological and Molecular Plant Pathology (2001) 59, 165±187 doi:10.1006/pmpp.2001.0355, available online at http://www.idealibrary.com on MINI-REVIEW The phylogeny of plant and animal pathogens in the Ascomycota MARY L. BERBEE* Department of Botany, University of British Columbia, 6270 University Blvd, Vancouver, BC V6T 1Z4, Canada (Accepted for publication August 2001) What makes a fungus pathogenic? In this review, phylogenetic inference is used to speculate on the evolution of plant and animal pathogens in the fungal Phylum Ascomycota. A phylogeny is presented using 297 18S ribosomal DNA sequences from GenBank and it is shown that most known plant pathogens are concentrated in four classes in the Ascomycota. Animal pathogens are also concentrated, but in two ascomycete classes that contain few, if any, plant pathogens. Rather than appearing as a constant character of a class, the ability to cause disease in plants and animals was gained and lost repeatedly. The genes that code for some traits involved in pathogenicity or virulence have been cloned and characterized, and so the evolutionary relationships of a few of the genes for enzymes and toxins known to play roles in diseases were explored. In general, these genes are too narrowly distributed and too recent in origin to explain the broad patterns of origin of pathogens. Co-evolution could potentially be part of an explanation for phylogenetic patterns of pathogenesis. Robust phylogenies not only of the fungi, but also of host plants and animals are becoming available, allowing for critical analysis of the nature of co-evolutionary warfare. Host animals, particularly human hosts have had little obvious eect on fungal evolution and most cases of fungal disease in humans appear to represent an evolutionary dead end for the fungus. -

Genetic Diversity of Barley Foliar Fungal Pathogens
agronomy Review Genetic Diversity of Barley Foliar Fungal Pathogens Arzu Çelik O˘guz* and Aziz Karakaya Department of Plant Protection, Faculty of Agriculture, Ankara University, Dı¸skapı,Ankara 06110, Turkey; [email protected] * Correspondence: [email protected] Abstract: Powdery mildew, net blotch, scald, spot blotch, barley stripe, and leaf rust are important foliar fungal pathogens of barley. Fungal leaf pathogens negatively affect the yield and quality in barley plant. Virulence changes, which can occur in various ways, may render resistant plants to susceptible ones. Factors such as mutation, population size and random genetic drift, gene and genotype flow, reproduction and mating systems, selection imposed by major gene resistance, and quantitative resistance can affect the genetic diversity of the pathogenic fungi. The use of fungicide or disease-resistant barley genotypes is an effective method of disease control. However, the evolutionary potential of pathogens poses a risk to overcome resistance genes in the plant and to neutralize fungicide applications. Factors affecting the genetic diversity of the pathogen fungus may lead to the emergence of more virulent new pathotypes in the population. Understanding the factors affecting pathogen evolution, monitoring pathogen biology, and genetic diversity will help to develop effective control strategies. Keywords: barley; Hordeum vulgare; Blumeria graminis; Pyrenophora teres; Rhynchosporium commune; Cochliobolus sativus; Pyrenophora graminea; Puccinia hordei; genetic diversity Citation: Çelik O˘guz,A.; Karakaya, A. Genetic Diversity of Barley Foliar Fungal Pathogens. Agronomy 2021, 11, 1. Introduction 434. https://doi.org/10.3390/ Barley (Hordeum vulgare L.) is one of the most important cereal crops that has been agronomy11030434 grown for thousands of years since prehistoric times, and is used in animal feed, malt products, and the food industry. -

The Response of Some Spring Barley Cultivars Grown in Finland to Air-Borne Secondary Infection by Bipolaris Sorokiniana of Infe
JOURNAL OF AGRICULTURAL SCIENCE IN FINLAND Maataloustieteellinen Aikakauskirja Vol. 57: 97—105, 1985 The response of some spring barley cultivars grown in Finland to air-borne secondary infection by Bipolaris sorokiniana AARNE KURPPA Department of Plant Pathology, University of Helsinki * SF-00710 HELSINKI 71, Finland Abstract. Air-borne secondary inoculum of Bipolaris sorokiniana caused severe foliar dis- eases and yield losses in all 12 spring barley cultivars tested in greenhouses or in the field. For secondary infection to occur a high relative humidity was necessary. Yield losses due to foliar diseases reached a maximum of 43.4 % in greenhouse experiments and 27.8 % in the field. The mean losses were 20.3 % and 12.3 %, respectively. Early infection at the time of heading or shortly after it resulted in higher yield losses than did later infection, although the symptom expression was opposite. Spore inoculation or natural secondary infection by the spores from a diseased crop after heading always resulted in a high infection incidence in the grain. Infec- tion incidence as well as fungal invasion of the internal cell leyers of the grains varied signifi- cantly among barley cultivars. The most susceptible of those tested were cvs. Teemu, Paavo and Pomo, while the most resistant were Ingrid, Otra and Pirkka. Introduction increasingly common in barley in the cool climate in North-Western Europe (Jorgen- sorokiniana (Sacc. in Sorok.) Bipolaris sen 1974, Hewett 1975, Mäkelä 1975, Shoem. (syn. Helminlhosporium sativum Kurppa 1984). & Cochlio- Pamm., King Bakke), perfect state Conidia of the fungus are the main sources sativus & Kurib.) has a world-wide bolus (Ito of infection, and are able to survive at least as a major pathogen of cereals distribution two years in soil (Ledingham 1970). -

Resistance in Spring Wheat to the Various Diseases Caused By
Resistance in spring wheat to the various diseases caused by Cochliobolus sativus by Aftabuddin Ahmed A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Plant Pathology Montana State University © Copyright by Aftabuddin Ahmed (1989) Abstract: Sixteen spring wheat cultivate were tested for their resistance to the various diseases caused by Cochliobolus sativus (Ito and Kurib.) Drechsl. ex Dastur. Five Montana isolates and four Bangladesh isolates of the fungus were used in whole plant inoculations in the Plant Growth Center and in laboratory tests using detached leaves. Sources of resistance were detected and identified for different phases of the disease. Ten cultivars were resistant to root rot, eight cultivars were resistant to foliar spot blotch, and six cultivars were resistant to head blight or black point. A number of cultivars showed differential reaction to various phases of the disease, e.g., were resistant to root rot but were susceptible to foliar spot blotch. Six cultivars, namely Marberg, GP248, GP253, GP254 and GP255, were resistant to all phases of the disease. The isolates tested differed significantly in pathogenicity, but considerable shifting in ranking occurred between experiments. Isolates obtained from roots were able to attack foliage/heads and vice versa. The isolates from Bangladesh did not have a higher temperature requirement than the USA isolates. Some cultivars were resistant to all isolates from both Bangladesh and the USA. The maximum disease development for root rot, foliar and head blight/black point phases occurred at 30°C with a 72 hour exposure to moist conditions. The disease reactions on detached leaves were not consistent with those on intact leaves. -

Spot Blotch), Fusarium
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.30.228932; this version posted March 24, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Genetics of resistance to common root rot (spot blotch), Fusarium crown rot, and sharp eyespot in wheat Jun Su 1,†, Jiaojie Zhao 1,†, Shuqing Zhao 1,†, Mengyu Li 1, Shuyong Pang 1,4, Shisheng Chen 4, Feng Chen 5, Wenchao Zhen 3,*, Zhensheng Kang 2,*, Xiaodong Wang 1,* 1 State Key Laboratory of North China Crop Improvement and Regulation / College of Plant Protection, Hebei Agricultural University, Baoding, Hebei 071000, P. R. China; 2 State Key Laboratory of Crop Stress Biology for Arid Areas / College of Plant Protection, Northwest A&F University, Yangling, Shaanxi 712100, P. R. China; 3 College of Agronomy, Hebei Agricultural University, Baoding, Hebei 071000, P. R. China; 4 Institute of Advanced Agricultural Sciences, Peking University, Weifang, Shandong 261000, P. R. China; 5 National Key Laboratory of Wheat and MaiZe Crop Science / Agronomy College, Henan Agricultural University, ZhengZhou 450046, P. R. China; * Correspondence: Xiaodong Wang ([email protected]), Zhensheng Kang ([email protected]), and Wenchao Zhen ([email protected]) † These authors contributed equally to this work. Abstract: Due to soil changes, high density planting, and the use of straw-returning methods, wheat common root rot (spot blotch), Fusarium crown rot (FCR), and sharp eyespot (sheath blight) have become severe threats to global wheat production. -

Efficient Discovery of Single-Nucleotide Variations in Cochliobolus Sativus Vegetative Compatibility Groups by Ecotilling
iochemis t B try n & la P P Jawhar et al., J Plant Biochem Physiol 2018, 6:2 h f y o s Journal of Plant Biochemistry & l DOI: 10.4172/2329-9029.1000211 i o a l n o r g u y o J Physiology ISSN: 2329-9029 Research Article Open Access Efficient Discovery of Single-Nucleotide Variations in Cochliobolus sativus Vegetative Compatibility Groups by Ecotilling Jawhar M1*, Arabi MIE1, MirAli N1 and Till BJ2 1Department of Molecular Biology and Biotechnology, AECS, Damascus, Syria 2Plant Breeding and Genetics Laboratory Vienna, Austria *Corresponding author: Jawhar M, Department of Molecular Biology and Biotechnology, AECS, Damascus, Syria, Tel: +2138520; E-mail: [email protected] Received date: March 06, 2018; Accepted date: March 12, 2018; Published date: March 23, 2018 Copyright: © 2018 Jawhar M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Cochliobolus sativus, the causal agent of barley spot blotch disease, is able to undergo spontaneous, vegetative cell fusion. In this study, the utility of enzymatic mismatch cleavage for discovery of single-nucleotide variations in Syrian vegetative compatibility groups (VCGs) obtained from pairing complementary nit mutants of C. sativus isolates was investigated for the first time. Gene-specific primers were designed from the whole fungal genome for use in the Ecotilling assays on VCGs isolates. Differences of band patterns among the digested products of different isolates were clearly observed by agarose gel electrophoresis. -

Multi-Locus Phylogeny of Pleosporales: a Taxonomic, Ecological and Evolutionary Re-Evaluation
available online at www.studiesinmycology.org StudieS in Mycology 64: 85–102. 2009. doi:10.3114/sim.2009.64.04 Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation Y. Zhang1, C.L. Schoch2, J. Fournier3, P.W. Crous4, J. de Gruyter4, 5, J.H.C. Woudenberg4, K. Hirayama6, K. Tanaka6, S.B. Pointing1, J.W. Spatafora7 and K.D. Hyde8, 9* 1Division of Microbiology, School of Biological Sciences, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, P.R. China; 2National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, 45 Center Drive, MSC 6510, Bethesda, Maryland 20892-6510, U.S.A.; 3Las Muros, Rimont, Ariège, F 09420, France; 4CBS-KNAW Fungal Biodiversity Centre, P.O. Box 85167, 3508 AD, Utrecht, The Netherlands; 5Plant Protection Service, P.O. Box 9102, 6700 HC Wageningen, The Netherlands; 6Faculty of Agriculture & Life Sciences, Hirosaki University, Bunkyo-cho 3, Hirosaki, Aomori 036-8561, Japan; 7Department of Botany and Plant Pathology, Oregon State University, Corvallis, Oregon 93133, U.S.A.; 8School of Science, Mae Fah Luang University, Tasud, Muang, Chiang Rai 57100, Thailand; 9International Fungal Research & Development Centre, The Research Institute of Resource Insects, Chinese Academy of Forestry, Kunming, Yunnan, P.R. China 650034 *Correspondence: Kevin D. Hyde, [email protected] Abstract: Five loci, nucSSU, nucLSU rDNA, TEF1, RPB1 and RPB2, are used for analysing 129 pleosporalean taxa representing 59 genera and 15 families in the current classification ofPleosporales . The suborder Pleosporineae is emended to include four families, viz. Didymellaceae, Leptosphaeriaceae, Phaeosphaeriaceae and Pleosporaceae. In addition, two new families are introduced, i.e. -

Race Structure of Pyrenophora Tritici-Repentis Isolates Obtained from Wheat in South America
Plant Protection Science – 2002 Plant Protection Science – 2002 Vol. 38, Special Issue 2: 302–304 Race Structure of Pyrenophora tritici-repentis Isolates Obtained from Wheat in South America A. SHAUKAT* and L. J. FRANCL Department of Plant Pathology, North Dakota State University, Fargo, ND 58105, USA *E-mail: [email protected] Abstract Knowledge of genetic variation in a pathogen population contributes to breeding for disease resistance. The fungus Pyrenophora tritici-repentis, cause of tan spot of wheat, is an important foliar pathogen worldwide. Currently, eight races have been identified in the fungal population prevalent on wheat and alternative hosts. Races 1 through 6 have been observed in North America. However, the fungal population from South America has not been characterized as to race. In this study, 48 single-spore isolates of P. tritici-repentis, recovered from wheat, were obtained from Argentina (10), Brazil (23), and Uruguay (15). Isolates were tested by inoculating individually on 2-leaf stage seedlings of the wheat differentials Glenlea, Katepwa, ND495, 6B365, M-3, and Salamouni in the greenhouse. They were grouped into differ- ent races based on necrosis and chlorosis induction on the differentials. Isolates from Argentina were grouped into races 1 and 7; from Brazil into races 1 and 8; and, from Uruguay into races 1 and 2. Results indicate that P. tritici-repentis has a diverse population on wheat in South America. More isolates are under investigation to obtain a comprehensive virulence pattern of the pathogen population in South America. Wheat lines should be screened against all known races to achieve durable resistance in a cultivar release program. -
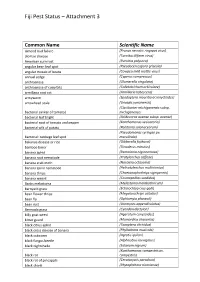
Fiji Pest Status – Attachment 3
Fiji Pest Status – Attachment 3 Common Name Scientific Name almond bud failure (Prunus necrotic ringspot virus) alomae disease (Taro bacilliform virus) American corn rust (Puccinia polysora) angular bean leaf spot (Pseudocercospora griseola) angular mosaic of beans (Cowpea mild mottle virus) annual sedge (Cyperus compressus) anthracnose (Glomerella cingulata) anthracnose of cucurbits (Colletotrichum orbiculare) armillaria root rot (Armillaria tabescens) armyworm (Spodoptera mauritia acronyctoides) arrowhead scale (Unaspis yanonensis) (Clavibacter michiganensis subsp. bacterial canker of tomato) michiganensis bacterial leaf blight (Acidovorax avenae subsp. avenae) bacterial spot of tomato and pepper (Xanthomonas vesicatoria) bacterial wilt of potato (Ralstonia solanacearum) (Pseudomonas syringae pv. bacterial: cabbage leaf spot maculicola) bakanae disease or rice (Gibberella fujikuroi) bamboo borer (Dinoderus minutus) banana aphid (Pentalonia nigronervosa) banana root nematode (Pratylenchus coffeae) banana scab moth (Nacoleia octasema) banana spiral nematode (Helicotylenchus multicinctus) banana thrips (Chaetanaphothrips signipennis) banana weevil (Cosmopolites sordidus) Banks melastoma (Melastoma malabathricum) barnyard grass (Echinochloa crus-galli) bean flower thrips (Megalurothrips usitatus) bean fly (Ophiomyia phaseoli) bean rust (Uromyces appendiculatus) Bermuda grass (Cynodon dactylon) billy goat weed (Ageratum conyzoides) bitter gourd (Momordica charantia) black citrus aphid (Toxoptera citricidus) black cross disease of banana (Phyllachora -

6Mycelial Growth Variation and Compatibility of the Selected Isolates of Bipolaris Sorokiniana (Sacc.) Shoemaker
Dhaka Univ. J. Biol. Sci. 28(2): 195-209, 2019 (July) 6MYCELIAL GROWTH VARIATION AND COMPATIBILITY OF THE SELECTED ISOLATES OF BIPOLARIS SOROKINIANA (SACC.) SHOEMAKER 1 2 MST. SELINA MOMTAZ , SHAMIM SHAMSI* AND TAPAN KUMAR DEY Department of Botany, University of Dhaka, Dhaka-1000, Bangladesh Key words: Cultural variation, Mycelial compatibility, Bipolaris sorokiniana, Leaf blight, Wheat Abstract Bipolaris sorokiniana (Sacc.) Shoemaker is a phytopathogenic fungus, causal agent of leaf blight disease of wheat. This fungus is difficult to control because of its immense cultural, morphological, physiological, pathogenic and genetic variability. The aim of this investigation was to study the cultural variability of the selected isolates of B. sorokiniana by means of mycelial compatibility. One hundred fifty isolates of B. sorokiniana from eight districts (Dhaka, Gazipur, Dinajpur, Joypurhat, Pabna, Sirajgonj, Kushtia and Chuadanga) of Bangladesh were studied and cultural variation was characterized on the basis of colony color and texture on PDA medium. They were - Black-Mat (B-M), Black-Fluffy (B-F), Ash-Mat (A-M), Brownish Ash-Fluffy (Br. A-F), Blackish Ash-Mat (Bl.A- M), Whitish Ash-Mat (W. A-M), Greenish Ash-Fluffy (G. A-F) and Pinkish White-Mat (P.W-M). To compare and understand cultural variability, mycelial compatibility test was done among the cultural groups. Results revealed that selected isolates of different cultural groups showed incompatible reaction. Incompatibility was also noticed between the isolates of same district and different districts. Introduction Wheat (Triticum aestivum L.) is the second most important cereal crop next to rice in Bangladesh. During 2016 - 2017, total wheat production was 1.311 million tons from 0.415 million hectares of land. -

Root and Crown Rots of Smal
PP-785 (Revised,) August 1999 Robert W. Stack, Professor Department of Plant Pathology Marcia McMullen, Extension Plant Pathologist Introduction Root/Crown Rots of Spring Wheat and Barley - Common Root Rot - Take-All - Fusarium Crown or Foot Rot - Other Root and Crown Rots of Spring Wheat and Barley Root and Crown Rots of Winter Wheat - Strawbreaker and Sharp Eyespot - Snow Molds of Winter Wheat Seedling Blights of Wheat, Winter Wheat, and Barley Root/Crown Rots of Oats Root rots of wheat and other small grains have plagued North Dakota farmers ever since the earliest farming days. The first station botanist at the North Dakota Agricultural College was Professor H.L. Bolley, a plant pathologist. In the early years of this century, Bolley identified root rot of wheat as an important problem and the underlying cause of what was then called "wheat-sick soil." Bolley campaigned across the state for farmers to practice rotation and use good seed. He appeared at county fairs and other gatherings and had posters placed in store windows encouraging farmers to recognize the importance of root rot. Two of his posters are reproduced in Figure 5. Their recommendations are still valid some 80 years later! Figure 5. Posters by Prof. H.L. Bolley from 1909 onward to publicize the importance of common root rot. The recommendations are still valid today. (21KB b&w image) Root rot damage is seldom recognized until severe damage or death of the crop is seen, because the action of root rot fungi is at or below ground level. Root rot damage is often mistaken for drought injury, soil problems or weather damage.