The Fe Protein: an Unsung Hero of Nitrogenase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Use of Mathematical Modeling and Other Biophysical Methods For
USE OF MATHEMATICAL MODELING AND OTHER BIOPHYSICAL METHODS FOR INSIGHTS INTO IRON-RELATED PHENOMENA OF BIOLOGICAL SYSTEMS A Dissertation by JOSHUA D. WOFFORD Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Paul A. Lindahl Committee Members, David P. Barondeau Simon W. North Vishal M. Gohil Head of Department, Simon W. North December 2018 Major Subject: Chemistry Copyright 2018 Joshua D. Wofford ABSTRACT Iron is a crucial nutrient in most living systems. It forms the active centers of many proteins that are critical for many cellular functions, either by themselves or as Fe-S clusters and hemes. However, Fe is potentially toxic to the cell in high concentrations and must be tightly regulated. There has been much work into understanding various pieces of Fe trafficking and regulation, but integrating all of this information into a coherent model has proven difficult. Past research has focused on different Fe species, including cytosolic labile Fe or mitochondrial Fe-S clusters, as being the main regulator of Fe trafficking in yeast. Our initial modeling efforts demonstrate that both cytosolic Fe and mitochondrial ISC assembly are required for proper regulation. More recent modeling efforts involved a more rigorous multi- tiered approach. Model simulations were optimized against experimental results involving respiring wild-type and Mrs3/4-deleted yeast. Simulations from both modeling studies suggest that mitochondria possess a “respiratory shield” that prevents a vicious cycle of nanoparticle formation, ISC loss, and subsequent loading of mitochondria with iron. -
Nitroxide-Mediated Polymerization
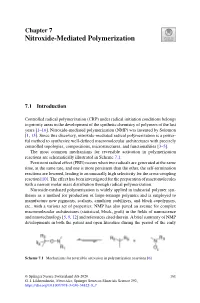
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

Characterization of a Microsomal Retinol Dehydrogenase Gene from Amphioxus: Retinoid Metabolism Before Vertebrates
Chemico-Biological Interactions 130–132 (2001) 359–370 www.elsevier.com/locate/chembiont Characterization of a microsomal retinol dehydrogenase gene from amphioxus: retinoid metabolism before vertebrates Diana Dalfo´, Cristian Can˜estro, Ricard Albalat, Roser Gonza`lez-Duarte * Departament de Gene`tica, Facultat de Biologia, Uni6ersitat de Barcelona, A6. Diagonal, 645, E-08028, Barcelona, Spain Abstract Amphioxus, a member of the subphylum Cephalochordata, is thought to be the closest living relative to vertebrates. Although these animals have a vertebrate-like response to retinoic acid, the pathway of retinoid metabolism remains unknown. Two different enzyme systems — the short chain dehydrogenase/reductases and the cytosolic medium-chain alcohol dehydrogenases (ADHs) — have been postulated in vertebrates. Nevertheless, recent data show that the vertebrate-ADH1 and ADH4 retinol-active forms originated after the divergence of cephalochordates and vertebrates. Moreover, no data has been gathered in support of medium-chain retinol active forms in amphioxus. Then, if the cytosolic ADH system is absent and these animals use retinol, the microsomal retinol dehydrogenases could be involved in retinol oxidation. We have identified the genomic region and cDNA of an amphioxus Rdh gene as a preliminary step for functional characterization. Besides, phyloge- netic analysis supports the ancestral position of amphioxus Rdh in relation to the vertebrate forms. © 2001 Elsevier Science Ireland Ltd. All rights reserved. Keywords: Retinol dehydrogenase; Retinoid metabolism; Amphioxus * Corresponding author. Fax: +34-93-4110969. E-mail address: [email protected] (R. Gonza`lez-Duarte). 0009-2797/01/$ - see front matter © 2001 Elsevier Science Ireland Ltd. All rights reserved. PII: S0009-2797(00)00261-1 360 D. -

Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase
Utah State University DigitalCommons@USU All Graduate Theses and Dissertations Graduate Studies 8-2018 Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase Rhesa N. Ledbetter Utah State University Follow this and additional works at: https://digitalcommons.usu.edu/etd Part of the Biochemistry Commons Recommended Citation Ledbetter, Rhesa N., "Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase" (2018). All Graduate Theses and Dissertations. 7197. https://digitalcommons.usu.edu/etd/7197 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@USU. It has been accepted for inclusion in All Graduate Theses and Dissertations by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. ELECTRON FLOW AND MANAGEMENT IN LIVING SYSTEMS: ADVANCING UNDERSTANDING OF ELECTRON TRANSFER TO NITROGENASE by Rhesa N. Ledbetter A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biochemistry Approved: ______________________ ______________________ Lance C. Seefeldt, Ph.D. Scott A. Ensign, Ph.D. Biochemistry Biochemistry Major Professor Committee Member ______________________ ______________________ Bruce Bugbee, Ph.D. Sean J. Johnson, Ph.D. Plant Physiology Biochemistry Committee Member Committee Member ______________________ ______________________ Nicholas E. Dickenson, Ph.D. Mark R. McLellan, Ph.D. Biochemistry Vice President for Research and Committee Member Dean of the School of Graduate Studies UTAH STATE UNIVERSITY Logan, Utah 2018 ii Copyright © Rhesa N. Ledbetter 2018 All Rights Reserved iii ABSTRACT Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase by Rhesa N. -

Methionine Sulfoxide Reductase a Is a Stereospecific Methionine Oxidase
Methionine sulfoxide reductase A is a stereospecific methionine oxidase Jung Chae Lim, Zheng You, Geumsoo Kim, and Rodney L. Levine1 Laboratory of Biochemistry, National Heart, Lung, and Blood Institute, Bethesda, MD 20892-8012 Edited by Irwin Fridovich, Duke University Medical Center, Durham, NC, and approved May 10, 2011 (received for review February 10, 2011) Methionine sulfoxide reductase A (MsrA) catalyzes the reduction Results of methionine sulfoxide to methionine and is specific for the S epi- Stoichiometry. Branlant and coworkers have studied in careful mer of methionine sulfoxide. The enzyme participates in defense detail the mechanism of the MsrA reaction in bacteria (17, 18). against oxidative stresses by reducing methionine sulfoxide resi- In the absence of reducing agents, each molecule of MsrA dues in proteins back to methionine. Because oxidation of methio- reduces two molecules of MetO. Reduction of the first MetO nine residues is reversible, this covalent modification could also generates a sulfenic acid at the active site cysteine, and it is function as a mechanism for cellular regulation, provided there reduced back to the thiol by a fast reaction, which generates a exists a stereospecific methionine oxidase. We show that MsrA disulfide bond in the resolving domain of the protein. The second itself is a stereospecific methionine oxidase, producing S-methio- MetO is then reduced and again generates a sulfenic acid at the nine sulfoxide as its product. MsrA catalyzes its own autooxidation active site. Because the resolving domain cysteines have already as well as oxidation of free methionine and methionine residues formed a disulfide, no further reaction forms. -

Electrochemical and Structural Characterization of Azotobacter Vinelandii Flavodoxin II
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II Helen M. Segal,1 Thomas Spatzal,1 Michael G. Hill,2 Andrew K. Udit,2 and Douglas C. Rees 1* 1Division of Chemistry and Chemical Engineering, Howard Hughes Medical Institute, California Institute of Technology, Pasadena, California 91125 2Division of Chemistry, Occidental College, Los Angeles, California 90041 Received 1 June 2017; Accepted 10 July 2017 DOI: 10.1002/pro.3236 Published online 14 July 2017 proteinscience.org Abstract: Azotobacter vinelandii flavodoxin II serves as a physiological reductant of nitrogenase, the enzyme system mediating biological nitrogen fixation. Wildtype A. vinelandii flavodoxin II was electrochemically and crystallographically characterized to better understand the molecular basis for this functional role. The redox properties were monitored on surfactant-modified basal plane graphite electrodes, with two distinct redox couples measured by cyclic voltammetry correspond- ing to reduction potentials of 2483 6 1 mV and 2187 6 9 mV (vs. NHE) in 50 mM potassium phos- phate, 150 mM NaCl, pH 7.5. These redox potentials were assigned as the semiquinone/ hydroquinone couple and the quinone/semiquinone couple, respectively. This study constitutes one of the first applications of surfactant-modified basal plane graphite electrodes to characterize the redox properties of a flavodoxin, thus providing a novel electrochemical method to study this class of protein. The X-ray crystal structure of the flavodoxin purified from A. vinelandii was solved at 1.17 A˚ resolution. With this structure, the native nitrogenase electron transfer proteins have all been structurally characterized. -

Genome-Wide Identification of Azospirillum Brasilense Sp245
Koul et al. BMC Genomics (2020) 21:821 https://doi.org/10.1186/s12864-020-07212-7 RESEARCH ARTICLE Open Access Genome-wide identification of Azospirillum brasilense Sp245 small RNAs responsive to nitrogen starvation and likely involvement in plant-microbe interactions Vatsala Koul1,2†, Divya Srivastava2†, Pushplata Prasad Singh2* and Mandira Kochar2* Abstract Background: Small RNAs (sRNAs) are non-coding RNAs known to regulate various biological functions such as stress adaptation, metabolism, virulence as well as pathogenicity across a wide range of bacteria, mainly by controlling mRNA stabilization or regulating translation. Identification and functional characterization of sRNAs has been carried out in various plant growth-promoting bacteria and they have been shown to help the cells cope up with environmental stress. No study has been carried out to uncover these regulatory molecules in the diazotrophic alpha-proteobacterium Azospirillum brasilense Sp245 to date. Results: Expression-based sRNA identification (RNA-seq) revealed the first list of ~ 468 sRNA candidate genes in A. brasilense Sp245 that were differentially expressed in nitrogen starvation versus non-starved conditions. In parallel, in silico tools also identified 2 of the above as candidate sRNAs. Altogether, putative candidates were stringently curated from RNA-seq data based on known sRNA parameters (size, location, secondary structure, and abundance). In total, ~ 59 significantly expressed sRNAs were identified in this study of which 53 are potentially novel sRNAs as they have no Rfam and BSRD homologs. Sixteen sRNAs were randomly selected and validated for differential expression, which largely was found to be in congruence with the RNA-seq data. Conclusions: Differential expression of 468 A. -

Respiration in Azotobacter Vinelandii and Its Relationship with the Synthesis of Biopolymers
Electronic Journal of Biotechnology 48 (2020) 36–45 Contents lists available at ScienceDirect Electronic Journal of Biotechnology Review Respiration in Azotobacter vinelandii and its relationship with the synthesis of biopolymers Tania Castillo a,AndrésGarcíab, Claudio Padilla-Córdova c, Alvaro Díaz-Barrera c,CarlosPeñaa,⁎ a Departamento de Ingeniería Celular y Biocatálisis, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Apdo. Post. 510-3, Cuernavaca 62250, Morelos, Mexico b Laboratorio de Biotecnología Ambiental, Centro de Investigación en Biotecnología, Universidad Autónoma del Estado de Morelos, Av. Universidad 1001, Col. Chamilpa, Cuernavaca, Morelos 62209, Mexico c Escuela de Ingeniería Bioquímica, Facultad de Ingeniería, Pontificia Universidad Católica de Valparaíso, Av. Brasil 2147, Casilla 4059, Valparaíso, Chile article info abstract Article history: Azotobacter vinelandii is a gram-negative soil bacterium that produces two biopolymers of biotechnological Received 20 April 2020 interest, alginate and poly(3-hydroxybutyrate), and it has been widely studied because of its capability to fix Accepted 27 August 2020 nitrogen even in the presence of oxygen. This bacterium is characterized by its high respiration rates, which Available online 4 September 2020 are almost 10-fold higher than those of Escherichia coli and are a disadvantage for fermentation processes. On the other hand, several works have demonstrated that adequate control of the oxygen supply in A. vinelandii Keywords: cultivations determines the yields and physicochemical characteristics of alginate and poly(3-hydroxybutyrate). poly(3-hydroxybutyrate) Azotobacter vinelandii Here, we summarize a review of the characteristics of A. vinelandii related to its respiration systems, as well as Alginate some of the most important findings on the oxygen consumption rates as a function of the cultivation Biopolymers parameters and biopolymer production. -

Diversity of Free-Living Nitrogen Fixing Bacteria in the Badlands of South Dakota Bibha Dahal South Dakota State University
South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange Theses and Dissertations 2016 Diversity of Free-living Nitrogen Fixing Bacteria in the Badlands of South Dakota Bibha Dahal South Dakota State University Follow this and additional works at: http://openprairie.sdstate.edu/etd Part of the Bacteriology Commons, and the Environmental Microbiology and Microbial Ecology Commons Recommended Citation Dahal, Bibha, "Diversity of Free-living Nitrogen Fixing Bacteria in the Badlands of South Dakota" (2016). Theses and Dissertations. 688. http://openprairie.sdstate.edu/etd/688 This Thesis - Open Access is brought to you for free and open access by Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. For more information, please contact [email protected]. DIVERSITY OF FREE-LIVING NITROGEN FIXING BACTERIA IN THE BADLANDS OF SOUTH DAKOTA BY BIBHA DAHAL A thesis submitted in partial fulfillment of the requirements for the Master of Science Major in Biological Sciences Specialization in Microbiology South Dakota State University 2016 iii ACKNOWLEDGEMENTS “Always aim for the moon, even if you miss, you’ll land among the stars”.- W. Clement Stone I would like to express my profuse gratitude and heartfelt appreciation to my advisor Dr. Volker Brӧzel for providing me a rewarding place to foster my career as a scientist. I am thankful for his implicit encouragement, guidance, and support throughout my research. This research would not be successful without his guidance and inspiration. -

A Brief Guide to Enzyme Classification and Nomenclature Rev AM
A Brief Guide to Enzyme Nomenclature and Classification Keith Tipton and Andrew McDonald 1) Introduction NC-IUBMB Enzyme List, or, to give it its full title, “Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes by the Reactions they Catalyse,1 is a functional system, based solely on the substrates transformed and products formed by an enzyme. The basic layout of the classification for each enzyme is described below with some indication of the guidelines followed. More detailed rules for enzyme nomenclature and classification are available online.2 Further details of the principles governing the nomenclature of individual enzyme classes are given in the following sections. 2. Basic Concepts 2.1. EC numbers Enzymes are identified by EC (Enzyme Commission) numbers. These are also valuable for relating the information to other databases. They were divided into 6 major classes according to the type of reaction catalysed and a seventh, the translocases, was added in 2018.3 These are shown in Table 1. Table 1. Enzyme classes Name Reaction catalysed 1 Oxidoreductases *AH2 + B = A +BH2 2 Transferases AX + B = BX + A 3 Hydrolases A-B + H2O = AH + BOH 4 Lyases A=B + X-Y = A-B ç ç X Y 5 Isomerases A = B 6 LiGases †A + B + NTP = A-B + NDP + P (or NMP + PP) 7 Translocases AX + B çç = A + X + ççB (side 1) (side 2) *Where nicotinamide-adenine dinucleotides are the acceptors, NAD+ and NADH + H+ are used, by convention. †NTP = nucleoside triphosphate. The EC number is made up of four components separated by full stops. -

Phylogeny of Nitrogenase Structural and Assembly Components Reveals New Insights Into the Origin and Distribution of Nitrogen Fixation Across Bacteria and Archaea
microorganisms Article Phylogeny of Nitrogenase Structural and Assembly Components Reveals New Insights into the Origin and Distribution of Nitrogen Fixation across Bacteria and Archaea Amrit Koirala 1 and Volker S. Brözel 1,2,* 1 Department of Biology and Microbiology, South Dakota State University, Brookings, SD 57006, USA; [email protected] 2 Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria 0004, South Africa * Correspondence: [email protected]; Tel.: +1-605-688-6144 Abstract: The phylogeny of nitrogenase has only been analyzed using the structural proteins NifHDK. As nifHDKENB has been established as the minimum number of genes necessary for in silico predic- tion of diazotrophy, we present an updated phylogeny of diazotrophs using both structural (NifHDK) and cofactor assembly proteins (NifENB). Annotated Nif sequences were obtained from InterPro from 963 culture-derived genomes. Nif sequences were aligned individually and concatenated to form one NifHDKENB sequence. Phylogenies obtained using PhyML, FastTree, RapidNJ, and ASTRAL from individuals and concatenated protein sequences were compared and analyzed. All six genes were found across the Actinobacteria, Aquificae, Bacteroidetes, Chlorobi, Chloroflexi, Cyanobacteria, Deferribacteres, Firmicutes, Fusobacteria, Nitrospira, Proteobacteria, PVC group, and Spirochaetes, as well as the Euryarchaeota. The phylogenies of individual Nif proteins were very similar to the overall NifHDKENB phylogeny, indicating the assembly proteins have evolved together. Our higher resolution database upheld the three cluster phylogeny, but revealed undocu- Citation: Koirala, A.; Brözel, V.S. mented horizontal gene transfers across phyla. Only 48% of the 325 genera containing all six nif genes Phylogeny of Nitrogenase Structural and Assembly Components Reveals are currently supported by biochemical evidence of diazotrophy. -

How Do Bacterial Cells Ensure That Metalloproteins Get the Correct Metal?
REVIEWS How do bacterial cells ensure that metalloproteins get the correct metal? Kevin J. Waldron and Nigel J. Robinson Abstract | Protein metal-coordination sites are richly varied and exquisitely attuned to their inorganic partners, yet many metalloproteins still select the wrong metals when presented with mixtures of elements. Cells have evolved elaborate mechanisms to scavenge for sufficient metal atoms to meet their needs and to adjust their needs to match supply. Metal sensors, transporters and stores have often been discovered as metal-resistance determinants, but it is emerging that they perform a broader role in microbial physiology: they allow cells to overcome inadequate protein metal affinities to populate large numbers of metalloproteins with the right metals. It has been estimated that one-quarter to one-third of all Both monovalent (cuprous) copper, which is expected proteins require metals, although the exploitation of ele- to predominate in a reducing cytosol, and trivalent ments varies from cell to cell and has probably altered over (ferric) iron, which is expected to predominate in an the aeons to match geochemistry1–3 (BOX 1). The propor- oxidizing periplasm, are also highly competitive, as are tions have been inferred from the numbers of homologues several non-essential metals, such as cadmium, mercury of known metalloproteins, and other deduced metal- and silver6 (BOX 2). How can a cell simultaneously contain binding motifs, encoded within sequenced genomes. A some proteins that require copper or zinc and others that large experimental estimate was generated using native require uncompetitive metals, such as magnesium or polyacrylamide-gel electrophoresis of extracts from iron- manganese? In a most simplistic model in which pro- rich Ferroplasma acidiphilum followed by the detection of teins pick elements from a cytosol in which all divalent metal in protein spots using inductively coupled plasma metals are present and abundant, all proteins would bind mass spectrometry4.