45758865004.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sex Differences in the Social Behavior of Juvenile Spider Monkeys (Ateles Geoffroyi) Michelle Amanda Rodrigues Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 Sex differences in the social behavior of juvenile spider monkeys (Ateles geoffroyi) Michelle Amanda Rodrigues Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Anthropology Commons Recommended Citation Rodrigues, Michelle Amanda, "Sex differences in the social behavior of juvenile spider monkeys (Ateles geoffroyi)" (2007). Retrospective Theses and Dissertations. 14829. https://lib.dr.iastate.edu/rtd/14829 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Sex differences in the social behavior of juvenile spider monkeys (Ateles geoffroyi) by Michelle Amanda Rodrigues A thesis submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF ARTS Major: Anthropology Program of Study Committee: Jill D. Pruetz, Major Professor W. Sue Fairbanks Maximilian Viatori Iowa State University Ames, Iowa 2007 Copyright © Michelle Amanda Rodrigues, 2007. All rights reserved. UMI Number: 1443144 UMI Microform 1443144 Copyright 2007 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. Box 1346 Ann Arbor, MI 48106-1346 ii For Travis, Goldie, Clydette, and Udi iii TABLE OF CONTENTS LIST OF TABLES vi LIST OF FIGURES vii ACKNOWLEDGMENTS viii ABSTRACT ix CHAPTER 1. -

Check List the Journal Of
12 3 1906 the journal of biodiversity data 19 June 2016 Check List NOTES ON GEOGRAPHIC DISTRIBUTION Check List 12(3): 1906, 19 June 2016 doi: http://dx.doi.org/10.15560/12.3.1906 ISSN 1809-127X © 2016 Check List and Authors A new population of the endangered Brachyteles arachnoides (É. Geoffroy, 1806) (Primates: Atelidae) in the state of Paraná, southern Brazil Bianca Ingberman1*, Nicholas Kaminski2, Roberto Fusco-Costa1 and Emygdio L.A. Monteiro-Filho1, 3 1 Instituto de Pesquisas Cananéia (IPeC), Wildlife Research Department, Rua Tristão Lobo 199, Centro, CEP 11990-000, Cananéia, SP, Brazil 2 Fundação Neotrópica do Brasil, Rua Dois de Outubro, 165, Bairro Recreio, CEP 79290-000, Bonito, MS, Brazil 3 Universidade Federal do Paraná, Centro Politécnico, Setor de Ciências Biológicas, Departamento de Zoologia, Laboratório de Biologia e Ecologia de Vertebrados, Caixa Postal 19020, Jardim das Américas, CEP 81531-990, Curitiba, PR, Brazil * Corresponding author. E-mail: [email protected] Abstract: The endangered southern muriqui or mono these states, but the distribution is much more restrict- [Brachyteles arachnoides (É. Geoffroy, 1806)], is a primate ed, fragmented and poorly known. Its presence in the endemic to the Atlantic Forest of Brazil. One known state of Paraná was suggested by Krieg (1939 apud Hill extant population is found at the southern limit of its 1962: 357), who stated that “… the range extends south- distribution, in the state of Paraná, where it is regionally wards beyond the Rio Ribeiro (i.e., Rio Ribeira de Igua- classified as Critically Endangered. Here, we report on pe) into the northern part of the state of Paraná...”. -

Diets of Howler Monkeys
Chapter 2 Diets of Howler Monkeys Pedro Américo D. Dias and Ariadna Rangel-Negrín Abstract Based on a bibliographical review, we examined the diets of howler mon- keys to compile a comprehensive overview of their food resources and document dietary diversity. Additionally, we analyzed the effects of rainfall, group size, and forest size on dietary variation. Howlers eat nearly all available plant parts in their habitats. Time dedicated to the consumption of different food types varies among species and populations, such that feeding behavior can range from high folivory to high frugivory. Overall, howlers were found to use at least 1,165 plant species, belonging to 479 genera and 111 families as food sources. Similarity in the use of plant taxa as food sources (assessed with the Jaccard index) is higher within than between howler species, although variation in similarity is higher within species. Rainfall patterns, group size, and forest size affect several dimensions of the dietary habits of howlers, such that, for instance, the degree of frugivory increases with increased rainfall and habitat size, but decreases with increasing group size in groups that live in more productive habitats. Moreover, the range of variation in dietary habits correlates positively with variation in rainfall, suggesting that some howler species are habitat generalists and have more variable diets, whereas others are habi- tat specialists and tend to concentrate their diets on certain plant parts. Our results highlight the high degree of dietary fl exibility demonstrated by the genus Alouatta and provide new insights for future research on howler foraging strategies. Resumen Con base en una revisión bibliográfi ca, examinamos las dietas de los monos aulladores para describir exhaustivamente sus recursos alimenticios y la diversidad de su dieta. -

Fission-Fusion Dynamics in Spider Monkeys in Belize
University of Calgary PRISM: University of Calgary's Digital Repository Graduate Studies The Vault: Electronic Theses and Dissertations 2016 Fission-Fusion Dynamics in Spider Monkeys in Belize Hartwell, Kayla Song Hartwell, K. S. (2016). Fission-Fusion Dynamics in Spider Monkeys in Belize (Unpublished doctoral thesis). University of Calgary, Calgary, AB. doi:10.11575/PRISM/26183 http://hdl.handle.net/11023/3497 doctoral thesis University of Calgary graduate students retain copyright ownership and moral rights for their thesis. You may use this material in any way that is permitted by the Copyright Act or through licensing that has been assigned to the document. For uses that are not allowable under copyright legislation or licensing, you are required to seek permission. Downloaded from PRISM: https://prism.ucalgary.ca UNIVERSITY OF CALGARY Fission-Fusion Dynamics in Spider Monkeys in Belize by Kayla Song Hartwell A THESIS SUBMITTED TO THE FACULTY OF GRADUATE STUDIES IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY GRADUATE PROGRAM IN ANTHROPOLOGY CALGARY, ALBERTA DECEMBER, 2016 © Kayla Song Hartwell 2016 Abstract Most diurnal primates live in cohesive social groups in which all or most members range in close proximity, but spider monkeys (Ateles) and chimpanzees (Pan) are known for their more fluid association patterns. These species have been traditionally described as living in fission-fusion societies, because they range in subgroups of frequently changing size and composition, in contrast with the more typical cohesive societies. In recent years the concept of fission-fusion dynamics has replaced the dichotomous fluid versus cohesive categorization, as it is now recognized that there is considerable variation in cohesiveness both within and between species. -

Parques Nacionais
National Parks Brazil BrasiParques Nacionails Brasil Parques Nacionais 2 3 4 5 National Parks Brazil BrasiParques Nacionails 6 7 O Brasil em sua imensidão abriga hoje 69 parques nacionais Brazil in its immensity today houses 69 national parks located situados nas cinco macro-regiões, protegendo no Norte áreas de in the five macro-regions, protecting the northern areas of florestas virgens e praticamente intocadas pelo homem, dunas e virgin forests – virtually untouched by man, dunes and rock pinturas rupestres no Nordeste, a exuberância de Mata Atlântica paintings in the Northeast, the exuberance of the Southeast no Sudeste, os Campos Gerais no Sul e uma flora e fauna do Atlantic Forest, Campos Gerais in the South and the exuberant exuberante do Cerrado no Centro-Oeste. Através desta publica- flora and fauna of the Cerrado in the Midwest. Through this ção a Localiza disponibiliza mais uma vez aos seus clientes e publication, Localiza makes available once more to its clients leitores a possibilidade de descoberta de exemplos bem suce- and readers the chance of discovering successful examples didos de manutenção da riqueza natural, legando às próximas of the maintenance of natural wealth, bequeathing to future gerações áreas de rara beleza. Juntas, elas compõem hoje um generations areas of outstanding beauty. Together they rico mosaico de preservação de nossa inigualável biodiversida- compose today a rich mosaic of conservation of our unique de, de nossa história e também nossa cultura. biodiversity, our history and our culture. Apoio Patrocínio Realização 8 9 Em 1876 o engenheiro abolicionista negro André Rebouças, foi precursor ao idealizar que o Brasil In 1876, the abolitionist engineer André Rebouças was a precursor when he idealized that Brazil destinasse parte de seu território para a criação de áreas protegidas com o intuito de salvaguardar would separate part of its territory to create protected areas with the intention to safeguard in a de forma sistemática, legal e organizada, aspectos importantes de nossos ecossistemas regionais. -

Fabiano R. Melo¹ and Luiz G. Dias²
Neotropical Primates 13(Suppl.), December 2005 19 MURIQUI POPULATIONS REPORTED IN THE LITERATURE OVER THE LAST 40 YEARS Fabiano R. Melo¹ and Luiz G. Dias² ¹Universidade Federal de Goiás, Campus Jataí, BR-364, Km 192, No. 3.800, Parque Industrial, Jataí 75801-615, Goiás, Brazil, e-mail: <[email protected]> ²Tropical Ecology, Assessment and Monitoring Network – Rio Doce, Universidade Federal de Minas Gerais, Av. Antônio Carlos 6627, Belo Horizonte 31270-901, MG, Brasil, e-mail: <[email protected]> Abstract Aguirre (O mono Brachyteles arachnoides (E. Geoff roy). Situação Atual da Espécie no Brasil. Acad. Brasil. Ciênc., Rio de Janei- ro, 1971) identifi ed 61 localities for the occurrence of the muriqui (Brachyteles arachnoides) in the states of Bahia, Espírito Santo, Minas Gerais, Rio de Janeiro, São Paulo and Paraná. He estimated a total population of 2,791 – 3,226 muriquis, con- trasting with a population of about 400,000 he reckoned would have existed in 1500. Accepting the position that there are two species, Aguirre’s (1971) data suggested a maximum of 996 individuals for the northern muriqui (Brachyteles hypoxan- thus) and about 2,230 for the southern muriqui (B. arachnoides). Current population estimates for the northern muriqui have indicated at least 864 individuals in the wild. Data available for the southern muriqui, suggest a minimum population of about 1,300. Th ese numbers combined approximate to the total population of 2,230 estimated by Aguirre (1971). Further population surveys for muriquis in the states São Paulo, Rio de Janeiro and Bahia are urgently needed, along with comparative studies on their basic ecology, diet and behavior in the diff erent-sized forest fragments and the more extensive forests. -
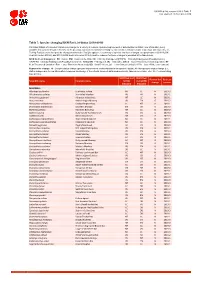
Table 7: Species Changing IUCN Red List Status (2018-2019)
IUCN Red List version 2019-3: Table 7 Last Updated: 10 December 2019 Table 7: Species changing IUCN Red List Status (2018-2019) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2018 (IUCN Red List version 2018-2) and 2019 (IUCN Red List version 2019-3) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered [CR(PE) - Critically Endangered (Possibly Extinct), CR(PEW) - Critically Endangered (Possibly Extinct in the Wild)], EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2018) List (2019) change version Category -

Genetic Structure in Two Northern Muriqui Populations (Brachyteles Hypoxanthus, Primates, Atelidae) As Inferred from Fecal DNA
Genetics and Molecular Biology, 31, 1, 166-171 (2008) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Short Communication Genetic structure in two northern muriqui populations (Brachyteles hypoxanthus, Primates, Atelidae) as inferred from fecal DNA Valéria Fagundes1, Marcela F. Paes1, Paulo B. Chaves1, Sérgio L. Mendes1, Carla de B. Possamai2,3, Jean P. Boubli4* and Karen B. Strier5 1Departamento de Ciências Biológicas, Universidade Federal do Espírito Santo, Vitória, ES, Brazil. 2RPPN Feliciano Miguel Abdala, Caratinga, MG, Brazil. 3Pontifícia Universidade Católica de Minas Gerais, Belo Horizonte, MG, Brazil. 4Conservation and Research for Endangered Species, Zoological Society of San Diego, San Diego, CA, USA. 5Department of Anthropology, University of Wisconsin-Madison, Wisconsin, USA. Abstract We assessed the genetic diversity of two northern muriqui (Brachyteles hypoxanthus Primata, Atelidae) populations, the Feliciano Miguel Abdala population (FMA, n = 108) in the Brazilian state of Minas Gerais (19°44’ S, 41°49’ W) and the Santa Maria de Jetibá population (SMJ, n = 18) in the Brazilian state of Espírito Santo (20°01’ S, 40°44’ W). Fecal DNA was isolated and PCR-RFLP analysis used to analyze 2160 bp of mitochondrial DNA, made up of an 820 bp segment of the gene cytochrome c oxidase subunit 2 (cox2, EC 1.9.3.1), an 880 bp segment of the gene cytochrome b(cytb, EC 1.10.2.2) and 460 bp of the hypervariable segment of the mtDNA control region (HVRI). The cox2 and cytb sequences were monomorphic within and between populations whereas the HVRI revealed three different pop- ulation exclusive haplotypes, one unique to the SMJ population and two, present at similar frequencies, in the FMA population. -

Atlantic Forest Southeast Reserves
WHC Nomination Documentation File Name: 893.pdf UNESCO Region: LATIN AMERICA AND THE CARIBBEANS __________________________________________________________________________________________________ SITE NAME: Atlantic Forest Southeast Reserves DATE OF INSCRIPTION: 4th December 1999 STATE PARTY: BRAZIL CRITERIA: N(ii)(iii)(iv) DECISION OF THE WORLD HERITAGE COMMITTEE: Excerpt from the Report of the 23rd Session of the World Heritage Committee IUCN informed the Committee that the evaluation of this property has been undertaken based on the revised nomination submitted by the State Party in April 1999. The Atlantic Forest Southeast Reserves contain the best and largest remaining examples of Atlantic forest in the Southeast region of Brazil. The 25 protected areas that make up the site display the biological richness and evolutionary history of the few remaining areas of Atlantic forest of Southeast Brazil. The area is also exceptionally diverse with high numbers of rare and endemic species. With its "mountains to the sea" attitudinal gradient, its estuary, wild rivers, karst and numerous waterfalls, the site also has exceptional scenic values. The Committee decided to inscribe the site under natural criteria (ii), (iii) and (iv). It also recommended that the State Party should be encouraged to restore natural conditions in the Serra do Mar State Park, which potentially could be incorporated in the site. The Delegate of Morocco noted the values of the site but highlighted the challenges of the management of serial sites. The Delegate of Australia noted that management in serial sites is complex but can be done with careful strategic planning and an appropriate legal framework. BRIEF DESCRIPTIONS The Atlantic Forest Southeast Reserves in the states of Parana and Sao Paolo, contain some of the best and largest examples of Atlantic forest in Brazil. -

Muriqui Brochure
FIND OUT muriqui Protect one ABOUT of the 25 most MONKEYSPACE Showcase your company’s brand endangered to the world, while engaging your employees in an Olympian challenge. This little guy can species in help! This is the wooly spider monkey, also known as muriqui: the the world most endangered primate in the Americas. He’s endangered because his forest habitat has been cleared for logging and farming. However, since being shortlisted as mascot for the Rio 2016 Olympics, it looks like his time has come. And your company can help safeguard his future, to the Olympics and beyond. SAVING THE MURIQUI OF SERRA DO © Bart van Dorp BRIGADEIRO © Bart van Dorp is all that remains of the IRACAMBI.COM muriqui’s habitat in the [email protected] 4% Atlantic Rainforest of Ph: +55 3139560454 Minas Gerais. WAYS TO HELP MONKEYSPACE IN SERRA DO BRIGADEIRO © Paulo B. Chavez Partner with us by adopting a Monkeyspace, a forest plot to Serra do Brigadeiro State Park extend the area of protected habitat in the Serra do Brigadeiro, Ervalia home to the largest northern Iracambi population of muriqui. With three and a half years to go before the Olympics, we invite you to join us Rosario da in this exciting adventure to create Limeira more forest land. © Bart van Dorp Muriaé Engage your staff in the challenge, and together let’s make sure that ABOUT THE the muriqui continues to thrive in his forest, Monkeyspace. MURIQUI (WOOLY SPIDER MONKEY) Also known as Mono Carvoeiro (Brachyteles will buy 50 hectares of forest land and hypoxanthus), the muriqui is considered one of the most endangered 25 species place it under permanent protection. -

Executive Summary of the National Action Plan For
EXECUTIVEEXECUTIVE SUMMARYSUMMARY OFOF THETHE NATIONALNATIONAL ACTIONACTION PLANPLAN FORFOR THETHE CONSERVATIONCONSERVATION OFOF MURIQUISMURIQUIS The mega-diverse country of Brazil is responsible for managing the largest natural patrimony in the world. More than 120,000 species of animals oc- cur throughout the country. Among these species, 627 are registered on the Official List of Brazilian Fauna Threatened with Extinction. The most affected Biome is the Atlantic Forest, within which 50% of the Critically Endangered mammalian species are primates endemic to this biome. Carla de Borba Possamai The Chico Mendes Institute is responsible for the development of strat- egies for the conservation of species of Brazilian fauna, evaluation of the conservation status of Brazilian fauna, publication of endangered species lists and red books and the development, implementation and monitoring of National Action Plans for the conservation of endangered species. Action plans are management tools for conservation of biodiversity and aligning strategies with different institutional stakeholders for the recovery and conservation of endangered species. The Joint Ordinance No. 316 of Septem- ber 9, 2009, was established as the legal framework to implement strategies. It indicates that action plans together with national lists of threatened species and red books, constitute one of the instruments of implementation of the National Biodiversity Policy (Decree 4.339/02). TAXANOMIC CLASSIFICATION Phylum: Chordata - Class: Mammalia - Order: Primates Family: Atelidae Gray, 1825 - Subfamily: Atelinae Gray, 1825 Genus: Brachyteles Spix, 1823 Species: Brachyteles arachnoides (E. Geoffroy, 1806) Brachyteles hypoxanthus (Kuhl, 1820) POPULAR NAMES Popular names include: Muriqui, Mono Carvoeiro, Mono, Miriqui, Buriqui, Buriquim, Mariquina or Muriquina. In English, they are also known as Woolly Spider Monkeys. -

A Review of the Studies on Colombian Primate Species
Mongabay.com Open Access Journal - Tropical Conservation Science Vol. 3(1):45-62, 2010 Research article Conservation of Colombian primates: an analysis of published research Stevenson, Pablo R.1; Guzmán, Diana C.1; Defler, Thomas R.2 1 Departamento de Ciencias Biológicas, Universidad de Los Andes, Cr. 1 no. 18ª-10, Bogotá, Colombia 2 Departamento de Biología, Universidad Nacional de Colombia, Cr. 45 no. 45-03, Bogotá. Corresponding author email: [email protected] Abstract In Colombia, there are approximately 27-31 primate species, including at least five endemic ones and a high proportion of threatened species. Differences in these primates’ distribution, abundance, ecology, and charisma, among many other things, have led to large variation in the amount and nature of investigations performed on the different species. Basic information on each native primate species is necessary to build adequate conservation plans; therefore, knowledge of the quantity and type of information available on each species can be helpful to identify possible research gaps. Based on publications from 1900 to 2008 on 25 primate species present in Colombia, we evaluated primate research in this country in terms of quantity, type, and topics of investigation. Additionally, we comparatively assessed the role of Colombian primatology within all scientific production on primate species present in this country. Our analyses indicate that in Colombia, primate research has developed mainly in the field-work area, with studies focused primarily on ecology and behavior. Investigations of topics such as karyology, anatomy, and physiology are very limited, and molecular biology is understudied compared to research on this subject in other countries.