Polymorphisms of Cytochrome P450 Genes in Three Ethnic Groups From
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification and Developmental Expression of the Full Complement Of
Goldstone et al. BMC Genomics 2010, 11:643 http://www.biomedcentral.com/1471-2164/11/643 RESEARCH ARTICLE Open Access Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish Jared V Goldstone1, Andrew G McArthur2, Akira Kubota1, Juliano Zanette1,3, Thiago Parente1,4, Maria E Jönsson1,5, David R Nelson6, John J Stegeman1* Abstract Background: Increasing use of zebrafish in drug discovery and mechanistic toxicology demands knowledge of cytochrome P450 (CYP) gene regulation and function. CYP enzymes catalyze oxidative transformation leading to activation or inactivation of many endogenous and exogenous chemicals, with consequences for normal physiology and disease processes. Many CYPs potentially have roles in developmental specification, and many chemicals that cause developmental abnormalities are substrates for CYPs. Here we identify and annotate the full suite of CYP genes in zebrafish, compare these to the human CYP gene complement, and determine the expression of CYP genes during normal development. Results: Zebrafish have a total of 94 CYP genes, distributed among 18 gene families found also in mammals. There are 32 genes in CYP families 5 to 51, most of which are direct orthologs of human CYPs that are involved in endogenous functions including synthesis or inactivation of regulatory molecules. The high degree of sequence similarity suggests conservation of enzyme activities for these CYPs, confirmed in reports for some steroidogenic enzymes (e.g. CYP19, aromatase; CYP11A, P450scc; CYP17, steroid 17a-hydroxylase), and the CYP26 retinoic acid hydroxylases. Complexity is much greater in gene families 1, 2, and 3, which include CYPs prominent in metabolism of drugs and pollutants, as well as of endogenous substrates. -

2016 SOT Enterocytes
Human Enterocytes: Isolation, Cryopreservation, Characterization, and Application in The Evaluation of Drug-Food Interactions Albert P. Li, Qian Yang, David Ho, Carol J. Loretz, Stephen N. Ring, Kirsten Amaral, Utkarsh Doshi, In Vitro ADMET Laboratories Inc., Columbia, MD and Malden, MA Introduction Results Why Enterocytes Successful Isolation and Comparison of Enterocytes and Hepatocytes in Drug Metabolizing Enzyme Activities (pmol/min/million enterocytes) Drug Gene Expression Substrate • Key cell type for oral bioavailability (as a Cryopreservation from Metabolizing Substrate Metabolite HE3005 HE3006 HE3007 HE3008 HE3009 HE3010 HE3011 HE3014 HE3015 HE3016 HE3020 HE3027 HE3029 Conc. (µM) function of permeability, metabolism, efflux) Multiple Donors Enzyme P450 Isoforms CYP2C9 Diclofenac 25 4-OH Diclofenac 1.68 0.59 0.91 0.46 1.18 1.21 0.03 0.44 2.50 2.05 0.31 2.02 0.86 First pass metabolism before the liver • CYP3A4/5 Midazolam 20 1-OH-Midazolam 2.67 0.13 0.99 0.87 0.72 0.46 0.09 0.40 2.55 0.99 0.49 0.68 0.59 Lot Gender Race Age (Years) -ΔCт -ΔCт Ratio of enterocyte to Gene name 2 enterocytes 2 hepatocytes 7-OH Coumarin UGT 7-OH Coumarin 100 8.38 2.30 3.08 1.80 4.32 2.56 1.01 3.55 7.33 5.71 5.83 3.68 6.55 • Target of drug-interactions with orally co- hepatocytes Glucuronide HE3005 M C 23 Sulfate administered substances CYP1A2 0.0000 0.1333 0.00 7-OH Coumarin 100 7-OH Coumarin Sulfate 8.72 2.04 4.04 1.79 7.78 3.32 1.70 2.66 5.23 4.13 1.84 2.69 3.65 HE3006 M C 47 Transferase CYP2B6 0.0024 1.0304 0.00 • Target of enterotoxicity of ingested -

Synonymous Single Nucleotide Polymorphisms in Human Cytochrome
DMD Fast Forward. Published on February 9, 2009 as doi:10.1124/dmd.108.026047 DMD #26047 TITLE PAGE: A BIOINFORMATICS APPROACH FOR THE PHENOTYPE PREDICTION OF NON- SYNONYMOUS SINGLE NUCLEOTIDE POLYMORPHISMS IN HUMAN CYTOCHROME P450S LIN-LIN WANG, YONG LI, SHU-FENG ZHOU Department of Nutrition and Food Hygiene, School of Public Health, Peking University, Beijing 100191, P. R. China (LL Wang & Y Li) Discipline of Chinese Medicine, School of Health Sciences, RMIT University, Bundoora, Victoria 3083, Australia (LL Wang & SF Zhou). 1 Copyright 2009 by the American Society for Pharmacology and Experimental Therapeutics. DMD #26047 RUNNING TITLE PAGE: a) Running title: Prediction of phenotype of human CYPs. b) Author for correspondence: A/Prof. Shu-Feng Zhou, MD, PhD Discipline of Chinese Medicine, School of Health Sciences, RMIT University, WHO Collaborating Center for Traditional Medicine, Bundoora, Victoria 3083, Australia. Tel: + 61 3 9925 7794; fax: +61 3 9925 7178. Email: [email protected] c) Number of text pages: 21 Number of tables: 10 Number of figures: 2 Number of references: 40 Number of words in Abstract: 249 Number of words in Introduction: 749 Number of words in Discussion: 1459 d) Non-standard abbreviations: CYP, cytochrome P450; nsSNP, non-synonymous single nucleotide polymorphism. 2 DMD #26047 ABSTRACT Non-synonymous single nucleotide polymorphisms (nsSNPs) in coding regions that can lead to amino acid changes may cause alteration of protein function and account for susceptivity to disease. Identification of deleterious nsSNPs from tolerant nsSNPs is important for characterizing the genetic basis of human disease, assessing individual susceptibility to disease, understanding the pathogenesis of disease, identifying molecular targets for drug treatment and conducting individualized pharmacotherapy. -

CYP2F1 (NM 000774) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC223097 CYP2F1 (NM_000774) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: CYP2F1 (NM_000774) Human Tagged ORF Clone Tag: Myc-DDK Symbol: CYP2F1 Synonyms: C2F1; CYP2F; CYPIIF1 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 4 CYP2F1 (NM_000774) Human Tagged ORF Clone – RC223097 ORF Nucleotide >RC223097 representing NM_000774 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGACAGCATAAGCACAGCCATCTTACTCCTGCTCCTGGCTCTCGTCTGTCTGCTCCTGACCCTAAGCT CAAGAGATAAGGGAAAGCTGCCTCCGGGACCCAGACCCCTCTCAATCCTGGGAAACCTGCTGCTGCTTTG CTCCCAAGACATGCTGACTTCTCTCACTAAGCTGAGCAAGGAGTATGGCTCCATGTACACAGTGCACCTG GGACCCAGGCGGGTGGTGGTCCTCAGCGGGTACCAAGCTGTGAAGGAGGCCCTGGTGGACCAGGGAGAGG AGTTTAGTGGCCGCGGTGACTACCCTGCCTTTTTCAACTTTACCAAGGGCAATGGCATCGCCTTCTCCAG TGGGGATCGATGGAAGGTCCTGAGACAGTTCTCTATCCAGATTCTACGGAATTTCGGGATGGGGAAGAGA AGCATTGAGGAGCGAATCCTAGAGGAGGGCAGCTTCCTGCTGGCGGAGCTGCGGAAAACTGAAGGCGAGC CCTTTGACCCCACGTTTGTGCTGAGTCGCTCAGTGTCCAACATTATCTGTTCCGTGCTCTTCGGCAGCCG CTTCGACTATGATGATGAGCGTCTGCTCACCATTATCCGCCTTATCAATGACAACTTCCAAATCATGAGC -

Metabolic Activation and Toxicological Evaluation of Polychlorinated Biphenyls in Drosophila Melanogaster T
www.nature.com/scientificreports OPEN Metabolic activation and toxicological evaluation of polychlorinated biphenyls in Drosophila melanogaster T. Idda1,7, C. Bonas1,7, J. Hofmann1, J. Bertram1, N. Quinete1,2, T. Schettgen1, K. Fietkau3, A. Esser1, M. B. Stope4, M. M. Leijs3, J. M. Baron3, T. Kraus1, A. Voigt5,6 & P. Ziegler1* Degradation of polychlorinated biphenyls (PCBs) is initiated by cytochrome P450 (CYP) enzymes and includes PCB oxidation to OH-metabolites, which often display a higher toxicity than their parental compounds. In search of an animal model refecting PCB metabolism and toxicity, we tested Drosophila melanogaster, a well-known model system for genetics and human disease. Feeding Drosophila with lower chlorinated (LC) PCB congeners 28, 52 or 101 resulted in the detection of a human-like pattern of respective OH-metabolites in fy lysates. Feeding fies high PCB 28 concentrations caused lethality. Thus we silenced selected CYPs via RNA interference and analyzed the efect on PCB 28-derived metabolite formation by assaying 3-OH-2′,4,4′-trichlorobiphenyl (3-OHCB 28) and 3′-OH-4′,4,6′-trichlorobiphenyl (3′-OHCB 28) in fy lysates. We identifed several drosophila CYPs (dCYPs) whose knockdown reduced PCB 28-derived OH-metabolites and suppressed PCB 28 induced lethality including dCYP1A2. Following in vitro analysis using a liver-like CYP-cocktail, containing human orthologues of dCYP1A2, we confrm human CYP1A2 as a PCB 28 metabolizing enzyme. PCB 28-induced mortality in fies was accompanied by locomotor impairment, a common phenotype of neurodegenerative disorders. Along this line, we show PCB 28-initiated caspase activation in diferentiated fy neurons. -

A Transcriptomic Approach to Elucidate the Physiological Significance Of
Madanayake et al. BMC Genomics 2013, 14:833 http://www.biomedcentral.com/1471-2164/14/833 RESEARCH ARTICLE Open Access A transcriptomic approach to elucidate the physiological significance of human cytochrome P450 2S1 in bronchial epithelial cells Thushara W Madanayake1, Ingrid E Lindquist2, Nicholas P Devitt2, Joann Mudge2 and Aaron M Rowland1* Abstract Background: Cytochrome P450 2S1 (CYP2S1) is an orphan P450 with an unknown biological function. Data from our laboratory and others suggest that CYP2S1 may have an important physiological role in modulating the synthesis and metabolism of bioactive lipids including prostaglandins and retinoids. CYP2S1 expression is elevated in multiple epithelial-derived cancers as well as in the chronic hyperproliferative disease psoriasis. Whether CYP2S1 expression in proliferative disease is protective, detrimental, or neutral to disease progression remains to be determined. Two human bronchial epithelial cells (BEAS-2B) were constructed to represent chronic depletion of CYP2S1 using short-hairpin RNA (shRNA) silencing directed toward the 3’UTR (759) and exon 3 (984) of the CYP2S1 gene and compared with a non-targeting shRNA control (SCRAM). Both CYP2S1 mRNA and protein were depleted by approximately 75% in stable cell lines derived from both targeted shRNA constructs (759 and 984). To elucidate the biological significance of CYP2S1, we analyzed transcriptome alterations in response to CYP2S1 depletion in human lung cells. Results: RNA-sequencing (RNA-seq) analysis was performed to compare the transcriptome of the control (SCRAM) and the CYP2S1-depleted (759) BEAS-2B cell lines. Transcriptomes of the replicates from the two cell lines were found to be distinct populations as determined using Principal Component Analysis and hierarchical clustering. -

Strong Correlation Between Air-Liquid Interface Cultures and in Vivo Transcriptomics of Nasal Brush Biopsy
Am J Physiol Lung Cell Mol Physiol 318: L1056–L1062, 2020. First published April 1, 2020; doi:10.1152/ajplung.00050.2020. RAPID REPORT Translational Physiology Strong correlation between air-liquid interface cultures and in vivo transcriptomics of nasal brush biopsy X Baishakhi Ghosh,1 Bongsoo Park,1 Debarshi Bhowmik,2 Kristine Nishida,3 Molly Lauver,3 Nirupama Putcha,3 Peisong Gao,4 Murugappan Ramanathan, Jr.,5 Nadia Hansel,3 Shyam Biswal,1 and Venkataramana K. Sidhaye1,3 1Department of Environmental Health and Engineering, Johns Hopkins Bloomberg School of Public Health, Baltimore, Maryland; 2Department of Biology, Johns Hopkins University, Baltimore, Maryland; 3Department of Pulmonary and Critical Care Medicine, Johns Hopkins School of Medicine, Baltimore, Maryland; 4Division of Allergy and Clinical Immunology, Johns Hopkins School of Medicine, Baltimore, Maryland; and 5Department of Otolaryngology-Head and Neck Surgery, Johns Hopkins School of Medicine, Baltimore, Maryland Submitted 18 February 2020; accepted in final form 24 March 2020 Ghosh B, Park B, Bhowmik D, Nishida K, Lauver M, Putcha N, RNA sequencing to determine the correlation between ALI- Gao P, Ramanathan M Jr, Hansel N, Biswal S, Sidhaye VK. cultured epithelial cells and epithelial cells obtained from nasal Strong correlation between air-liquid interface cultures and in vivo brushing and identify differences that may arise as a result of transcriptomics of nasal brush biopsy. Am J Physiol Lung Cell Mol redifferentiation. Physiol 318: L1056–L1062, 2020. First published April 1, 2020; doi:10.1152/ajplung.00050.2020.—Air-liquid interface (ALI) cultures METHODS are ex vivo models that are used extensively to study the epithelium of patients with chronic respiratory diseases. -

Conformational Changes in Binding of Substrates with Human Cytochrome P450 Enzymes
Book of oral abstracts 100 Conformational changes in binding of substrates with human cytochrome P450 enzymes F. Peter Guengerich, Clayton J. Wilkey, Michael J. Reddish, Sarah M. Glass, and Thanh T. N. Phan Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN, USA Introduction. Extensive evidence now exists that P450 enzymes can exist in multiple conformations, at least in the substrate-bound forms (e.g., crystallography). This multiplicity can be the result of either an induced fit mechanism or conformational selection (selective substrate binding to one of two or more equilibrating P450 conformations). Aims. Kinetic approaches can be used to distinguish between induced fit and conformational selection models. The same energy is involved in reaching the final state, regardless of the kinetic path. Methods. Stopped-flow absorbance and fluorescence measurements were made with recombinant human P450 enzymes. Analysis utilized kinetic modeling software (KinTek Explorer®). Results. P450 17A1 binding to its steroid ligands (pregnenolone and progesterone and the 17-hydroxy derivatives) is dominated by a conformational selection process, as judged by (a) decreasing rates of substrate binding as a function of substrate concentration, (b) opposite patterns of the dependence of binding rates as a function of varying concentrations of (i) substrate and (ii) enzyme, and (c) modeling of the data in KinTek Explorer. The inhibitory drugs orteronel and abiraterone bind P450 17A1 in multi-step processes, apparently in different ways. The dye Nile Red is also a substrate for P450 17A1 and its sequential binding to the enzyme can be resolved in fluorescence and absorbance changes. P450s 2C8, 2D6, 2E1, and 4A11 have also been analyzed with regard to substrate binding and utilize primarily conformational selection models, as revealed by analysis of binding rates vs. -

Expression of CYP2S1 in Human Hepatic Stellate Cells
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Letters 581 (2007) 781–786 Expression of CYP2S1 in human hepatic stellate cells Carylyn J. Mareka, Steven J. Tuckera, Matthew Korutha, Karen Wallacea,b, Matthew C. Wrighta,b,* a School of Medical Sciences, Institute of Medical Science, University of Aberdeen, Aberdeen, UK b Liver Faculty Research Group, School of Clinical and Laboratory Sciences, University of Newcastle, Newcastle, UK Received 22 November 2006; revised 16 January 2007; accepted 23 January 2007 Available online 2 February 2007 Edited by Laszlo Nagy the expression and accumulation of scarring extracellular Abstract Activated stellate cells are myofibroblast-like cells associated with the generation of fibrotic scaring in chronically fibrotic matrix protein [2]. It is currently thought an inhibition damaged liver. Gene chip analysis was performed on cultured fi- of fibrosis in liver diseases may be an effective approach to brotic stellate cells. Of the 51 human CYP genes known, 13 treating patients for which the cause is refractive to current CYP and 5 CYP reduction-related genes were detected with 4 treatments (e.g. in approx. 30% of hepatitis C infected CYPs (CYP1A1, CYP2E1, CY2S1 and CYP4F3) consistently patients) [2,3]. At present, there is no approved treatment for present in stellate cells isolated from three individuals. Quantita- fibrosis. tive RT-PCR indicated that CYP2S1 was a major expressed Inadvertent toxicity of drugs is often associated with a ‘‘met- CYP mRNA transcript. The presence of a CYP2A-related pro- abolic activation’’ by CYPs [1]. -

PDF (Supplementary Figures)
Supplementary Information Insights into an Efficient Light-driven Hybrid P450 BM3 Enzyme from Crystallographic, Spectroscopic and Biochemical Studies Jessica Spradlin,1 Diana Lee,1 Sruthi Mahadevan,1 Mavish Mahomed,2 Lawrence Tang,1 Quan Lam,1 Alexander Colbert,1 Oliver S. Shafaat,3 David Goodin,2 Marco Kloos,4 Mallory Kato,1 Lionel E. Cheruzel1* 1 San José State University, Department of Chemistry, One Washington Square, San José, CA 2 Department of Chemistry, One Shields Ave., University of California Davis, Davis, CA. 3 Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, CA. 4 Department of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Heidelberg, Germany. Table of Contents: Figure S1. Graphical representations of C-H---pi interactions with P382..……………………………p.S-2 Figure S2. Transient absorption traces for the various hybrid enzymes …………….………………...p.S-3 Figure S3. UV vis spectra for the substrate-free, NPG bound and reduced hybrid enzymes..….……..p.S-4 Figure S4. Michaelis-Menten saturation curves with 16-pNCA substrate………….…………...……..p.S-4 Figure S5. Partial sequence alignment of 40 human cytochrome P450 heme domains….……...……..p.S-5 S-1 Fig. S1. Graphical representations of C-H---π interactions between the photosensitizer and the P382 residue preserved during the motion of the 310 helix by 2.1 Å in the two molecules present in the asymmetric unit of the DMSO bound structure. S-2 Fig. S2. Transient absorption traces for the dF393A-1, dF393W-1 and sL407C-1 hybrid enzymes and the corresponding difference spectra for the substrate free (SF) and substrate bound (SB) forms (bottom panels). -
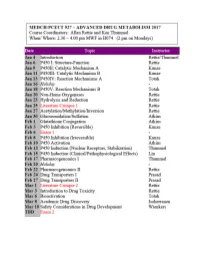
CYTOCHROME P450: Structure-Function
MEDCH 527 AER Jan. 4-6, 2017 CYTOCHROME P450: Structure-Function 1. General P450 Characteristics anD Taxonomy 2. Human P450s – Substrate anD Inhibitor Selectivities 3. Structure-Function Aspects of LiganD BinDing, P450 ReDuction anD Oxygen Activation References P450 Homepage -http:// http://drnelson.uthsc.edu/CytochromeP450.html Nelson DR et al., Pharmacogenetics. 1996 6(1):1-42: Nelson DR et al., Pharmacogenetics. 2004 14(1):1-18. Testa, B. The Biochemistry of Drug Metabolism: A 6 Part Series in Chem. BioDivers. (2006-2008). Sligar, SG. Glimpsing the critical intermediate in cytochrome P450 oxidations. Science. 2010 Nov 12;330(6006):924-5. Johnson EF, et al. Correlating structure and function of drug metabolizing enzymes:Progress and ongoing challenges. Drug Metab. Dispos. 42:9-22 (2014). Zientek MA and Youdim K, Reaction Phenotyping:Advances in experimental strategies used to characterize the contribution of drug metabolizing enzymes. Drug Metab. Dispos. 43:163-181 (2015). Almazroo et al., Drug Metabolism in the Liver. Clin. Liver Dis. 21:1-20 (2017). Absorption, Distribution, Metabolism and Excretion (ADME) of Orally Administered Drugs To reach their sites of action in the body, orally administered drugs must be absorbed from the small intestine, survive first pass metabolism - typically in the liver - before eventually being excreted, usually as drug metabolites in the bile and kidney. Therefore, clinically useful drugs must be able to cross an array of cell membranes, which are composed of a lipid bilayer. Drugs must exhibit an adequate degree of lipophilicity (logP of ~2-4) in order to able to dissolve into this lipoidal environment. Many drug metabolism processes render lipophilic drugs more water-soluble so as to facilitate excretion via the kidneys and bile. -

Pharmacogenetics of Cytochrome P450 and Its Application and Value in Drug Therapy – the Past, Present and Future
Pharmacogenetics of cytochrome P450 and its application and value in drug therapy – the past, present and future Magnus Ingelman-Sundberg Karolinska Institutet, Stockholm, Sweden The human genome x 3,120,000,000 nucleotides x 23,000 genes x >100 000 transcripts (!) x up to 100,000 aa differences between two proteomes x 10,000,000 SNPs in databases today The majority of the human genome is transcribed and has an unknown function RIKEN consortium Science 7 Sep 2005 Interindividual variability in drug action Ingelman-Sundberg, M., J Int Med 250: 186-200, 2001, CYP dependent metabolism of drugs (80 % of all phase I metabolism of drugs) Tolbutamide Beta blokers Warfarin Antidepressants Phenytoin CYP2C9* Diazepam Antipsychotics NSAID Citalopram Dextromethorphan CYP2D6* CYP2C19* Anti ulcer drugs Codeine CYP2E1 Clozapine Debrisoquine CYP1A2 Ropivacaine CYP2B6* Efavirenz Cyclophosphamide CYP3A4/5/7 Cyclosporin Taxol Tamoxifen Tacrolimus 40 % of the phase I Amprenavir Amiodarone metabolism is Cerivastatin carried out by Erythromycin polymorphic P450s Methadone Quinine (enzymes in Italics) Phenotypes and mutations PM, poor metabolizers; IM, intermediate met; EM, efficient met; UM, ultrarapid met Frequency Population Homozygous based dosing for • Stop codons • Heterozygous Two funct deleterious • Deletions alleles SNPs • Deleterious • Gene missense • Unstable duplication SNPs protein • Induction • Splice defects EM PM IM UM Enzyme activity/clearance The Home Page of the Human Cytochrome P450 (CYP) Allele Nomenclature Committee http://www.imm.ki.se/CYPalleles/ Webmaster: Sarah C Sim Editors: Magnus Ingelman-Sundberg, Ann K. Daly, Daniel W. Nebert Advisory Board: Jürgen Brockmöller, Michel Eichelbaum, Seymour Garte, Joyce A. Goldstein, Frank J. Gonzalez, Fred F. Kadlubar, Tetsuya Kamataki, Urs A.