Final Report: Describe Existing Populations And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Principles and Practices for Restoration of Ponderosa Pine and Dry Mixed
United States Department of Agriculture Principles and Practices for the Restoration of Ponderosa Pine and Dry Mixed-Conifer Forests of the Colorado Front Range Robert N. Addington, Gregory H. Aplet, Mike A. Battaglia, Jennifer S. Briggs, Peter M. Brown, Antony S. Cheng, Yvette Dickinson, Jonas A. Feinstein, Kristen A. Pelz, Claudia M. Regan, Jim Thinnes, Rick Truex, Paula J. Fornwalt, Benjamin Gannon, Chad W. Julian, Jeffrey L. Underhill, Brett Wolk Forest Rocky Mountain General Technical Report Service Research Station RMRS-GTR-373 January 2018 Addington, Robert N.; Aplet, Gregory H.; Battaglia, Mike A.; Briggs, Jennifer S.; Brown, Peter M.; Cheng, Antony S.; Dickinson, Yvette; Feinstein, Jonas A.; Pelz, Kristen A.; Regan, Claudia M.; Thinnes, Jim; Truex, Rick; Fornwalt, Paula J.; Gannon, Benjamin; Julian, Chad W.; Underhill, Jeffrey L.; Wolk, Brett. 2018. Principles and practices for the restoration of ponderosa pine and dry mixed-conifer forests of the Colorado Front Range. RMRS-GTR-373. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 121 p. Abstract Wildfires have become larger and more severe over the past several decades on Colorado’s Front Range, catalyzing greater investments in forest management intended to mitigate wildfire risks.The complex ecological, social, and political context of the Front Range, however, makes forest management challenging, especially where multiple management goals including forest restoration exist. In this report, we present a science-based framework for managers to develop place-based approaches to forest restoration of Front Range ponderosa pine and dry mixed-conifer forests. We first present ecological information describing how Front Range forest structure and composition are shaped at multiple scales by interactions among topography, natural disturbances such as fire, and forest developmental processes. -

Denudation History and Internal Structure of the Front Range and Wet Mountains, Colorado, Based on Apatite-Fission-Track Thermoc
NEW MEXICO BUREAU OF GEOLOGY & MINERAL RESOURCES, BULLETIN 160, 2004 41 Denudation history and internal structure of the Front Range and Wet Mountains, Colorado, based on apatitefissiontrack thermochronology 1 2 1Department of Earth and Environmental Science, New Mexico Institute of Mining and Technology, Socorro, NM 87801Shari A. Kelley and Charles E. Chapin 2New Mexico Bureau of Geology and Mineral Resources, New Mexico Institute of Mining and Technology, Socorro, NM 87801 Abstract An apatite fissiontrack (AFT) partial annealing zone (PAZ) that developed during Late Cretaceous time provides a structural datum for addressing questions concerning the timing and magnitude of denudation, as well as the structural style of Laramide deformation, in the Front Range and Wet Mountains of Colorado. AFT cooling ages are also used to estimate the magnitude and sense of dis placement across faults and to differentiate between exhumation and faultgenerated topography. AFT ages at low elevationX along the eastern margin of the southern Front Range between Golden and Colorado Springs are from 100 to 270 Ma, and the mean track lengths are short (10–12.5 µm). Old AFT ages (> 100 Ma) are also found along the western margin of the Front Range along the Elkhorn thrust fault. In contrast AFT ages of 45–75 Ma and relatively long mean track lengths (12.5–14 µm) are common in the interior of the range. The AFT ages generally decrease across northwesttrending faults toward the center of the range. The base of a fossil PAZ, which separates AFT cooling ages of 45– 70 Ma at low elevations from AFT ages > 100 Ma at higher elevations, is exposed on the south side of Pikes Peak, on Mt. -

To See the Hike Archive
Geographical Area Destination Trailhead Difficulty Distance El. Gain Dest'n Elev. Comments Allenspark 932 Trail Near Allenspark A 4 800 8580 Allenspark Miller Rock Riverside Dr/Hwy 7 TH A 6 700 8656 Allenspark Taylor and Big John Taylor Rd B 7 2300 9100 Peaks Allenspark House Rock Cabin Creek Rd A 6.6 1550 9613 Allenspark Meadow Mtn St Vrain Mtn TH C 7.4 3142 11632 Allenspark St Vrain Mtn St Vrain Mtn TH C 9.6 3672 12162 Big Thompson Canyon Sullivan Gulch Trail W of Waltonia Rd on Hwy A 2 941 8950 34 Big Thompson Canyon 34 Stone Mountain Round Mtn. TH B 8 2100 7900 Big Thompson Canyon 34 Mt Olympus Hwy 34 B 1.4 1438 8808 Big Thompson Canyon 34 Round (Sheep) Round Mtn. TH B 9 3106 8400 Mountain Big Thompson Canyon Hwy 34 Foothills Nature Trail Round Mtn TH EZ 2 413 6240 to CCC Shelter Bobcat Ridge Mahoney Park/Ginny Bobcat Ridge TH B 10 1500 7083 and DR trails Bobcat Ridge Bobcat Ridge High Bobcat Ridge TH B 9 2000 7000 Point Bobcat Ridge Ginny Trail to Valley Bobcat Ridge TH B 9 1604 7087 Loop Bobcat Ridge Ginny Trail via Bobcat Ridge TH B 9 1528 7090 Powerline Tr Boulder Chautauqua Park Royal Arch Chautauqua Trailhead by B 3.4 1358 7033 Rgr. Stn. Boulder County Open Space Mesa Trail NCAR Parking Area B 7 1600 6465 Boulder County Open Space Gregory Canyon Loop Gregory Canyon Rd TH B 3.4 1368 7327 Trail Boulder Open Space Heart Lake CR 149 to East Portal TH B 9 2000 9491 Boulder Open Space South Boulder Peak Boulder S. -

Colorado Fourteeners Checklist
Colorado Fourteeners Checklist Rank Mountain Peak Mountain Range Elevation Date Climbed 1 Mount Elbert Sawatch Range 14,440 ft 2 Mount Massive Sawatch Range 14,428 ft 3 Mount Harvard Sawatch Range 14,421 ft 4 Blanca Peak Sangre de Cristo Range 14,351 ft 5 La Plata Peak Sawatch Range 14,343 ft 6 Uncompahgre Peak San Juan Mountains 14,321 ft 7 Crestone Peak Sangre de Cristo Range 14,300 ft 8 Mount Lincoln Mosquito Range 14,293 ft 9 Castle Peak Elk Mountains 14,279 ft 10 Grays Peak Front Range 14,278 ft 11 Mount Antero Sawatch Range 14,276 ft 12 Torreys Peak Front Range 14,275 ft 13 Quandary Peak Mosquito Range 14,271 ft 14 Mount Evans Front Range 14,271 ft 15 Longs Peak Front Range 14,259 ft 16 Mount Wilson San Miguel Mountains 14,252 ft 17 Mount Shavano Sawatch Range 14,231 ft 18 Mount Princeton Sawatch Range 14,204 ft 19 Mount Belford Sawatch Range 14,203 ft 20 Crestone Needle Sangre de Cristo Range 14,203 ft 21 Mount Yale Sawatch Range 14,200 ft 22 Mount Bross Mosquito Range 14,178 ft 23 Kit Carson Mountain Sangre de Cristo Range 14,171 ft 24 Maroon Peak Elk Mountains 14,163 ft 25 Tabeguache Peak Sawatch Range 14,162 ft 26 Mount Oxford Collegiate Peaks 14,160 ft 27 Mount Sneffels Sneffels Range 14,158 ft 28 Mount Democrat Mosquito Range 14,155 ft 29 Capitol Peak Elk Mountains 14,137 ft 30 Pikes Peak Front Range 14,115 ft 31 Snowmass Mountain Elk Mountains 14,099 ft 32 Windom Peak Needle Mountains 14,093 ft 33 Mount Eolus San Juan Mountains 14,090 ft 34 Challenger Point Sangre de Cristo Range 14,087 ft 35 Mount Columbia Sawatch Range -
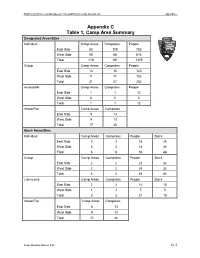
Appendix C Table 1, Camp Area Summary
Backcountry/Wilderness Management Plan and Environmental Assessment Appendix C Appendix C Table 1, Camp Area Summary Designated Areas/Sites Individual Camp Areas Campsites People East Side 60 109 763 West Side 59 88 616 Total 119 197 1379 Group Camp Areas Campsites People East Side 10 10 120 West Side 11 11 132 Total 21 21 252 Accessible Camp Areas Campsites People East Side 1 1 12 West Side 0 0 0 Total 1 1 12 Wood Fire Camp Areas Campsites East Side 8 13 West Side 9 13 Total 17 26 Stock Areas/Sites Individual Camp Areas Campsites People Stock East Side 3 3 18 24 West Side 3 3 18 24 Total 6 6 36 48 Group Camp Areas Campsites People Stock East Side 2 2 24 32 West Side 2 2 24 32 Total 4 4 48 64 Llama only Camp Areas Campsites People Stock East Side 2 2 14 10 West Side1175 Total 3 3 21 15 Wood Fire Camp Areas Campsites East Side 8 13 West Side 9 13 Total 17 26 Rocky Mountain National Park C-1 Backcountry/Wilderness Management Plan and Environmental Assessment Appendix C Crosscountry Areas Areas Parties People East Side 9 16 112 West Side 14 32 224 Total 23 48 336 Summer Totals for Designated, Stock and Crosscountry Areas Camp Areas Campsites/Parties People East Side 80 136 1004 West Side 84 131 969 Total 164 267 1973 Bivouac Areas Areas People East Side 11 88 West Side 0 0 Total 11 88 Winter Areas Areas Parties People East Side 32 136 1632 West Side 23 71 852 Total 55 207 2484 Rocky Mountain National Park C-2 Backcountry/Wilderness Management Plan and Environmental Assessment Appendix C Appendix C Table 2, Designated Camp Area/Sites Number -

Colorado Climate Center Sunset
Table of Contents Why Is the Park Range Colorado’s Snowfall Capital? . .1 Wolf Creek Pass 1NE Weather Station Closes. .4 Climate in Review . .5 October 2001 . .5 November 2001 . .6 Colorado December 2001 . .8 Climate Water Year in Review . .9 Winter 2001-2002 Why Is It So Windy in Huerfano County? . .10 Vol. 3, No. 1 The Cold-Land Processes Field Experiment: North-Central Colorado . .11 Cover Photo: Group of spruce and fi r trees in Routt National Forest near the Colorado-Wyoming Border in January near Roger A. Pielke, Sr. Colorado Climate Center sunset. Photo by Chris Professor and State Climatologist Department of Atmospheric Science Fort Collins, CO 80523-1371 Hiemstra, Department Nolan J. Doesken of Atmospheric Science, Research Associate Phone: (970) 491-8545 Colorado State University. Phone and fax: (970) 491-8293 Odilia Bliss, Technical Editor Colorado Climate publication (ISSN 1529-6059) is published four times per year, Winter, Spring, If you have a photo or slide that you Summer, and Fall. Subscription rates are $15.00 for four issues or $7.50 for a single issue. would like considered for the cover of Colorado Climate, please submit The Colorado Climate Center is supported by the Colorado Agricultural Experiment Station it to the address at right. Enclose a note describing the contents and through the College of Engineering. circumstances including loca- tion and date it was taken. Digital Production Staff: Clara Chaffi n and Tara Green, Colorado Climate Center photo graphs can also be considered. Barbara Dennis and Jeannine Kline, Publications and Printing Submit digital imagery via attached fi les to: [email protected]. -

Rocky Mountain National Park Lawn Lake Flood Interpretive Area (Elevation 8,640 Ft)
1 NCSS Conference 2001 Field Tour -- Colorado Rocky Mountains Wednesday, June 27, 2001 7:00 AM Depart Ft. Collins Marriott 8:30 Arrive Rocky Mountain National Park Lawn Lake Flood Interpretive Area (elevation 8,640 ft) 8:45 "Soil Survey of Rocky Mountain National Park" - Lee Neve, Soil Survey Project Leader, Natural Resources Conservation Service 9:00 "Correlation and Classification of the Soils" - Thomas Hahn, Soil Data Quality Specialist, MLRA Office 6, Natural Resources Conservation Service 9:15-9:30 "Interpretive Story of the Lawn Lake Flood" - Rocky Mountain National Park Interpretive Staff, National Park Service 10:00 Depart 10:45 Arrive Alpine Visitors Center (elevation 11,796 ft) 11:00 "Research Needs in the National Parks" - Pete Biggam, Soil Scientist, National Park Service 11:05 "Pedology and Biogeochemistry Research in Rocky Mountain National Park" - Dr. Eugene Kelly, Colorado State University 11:25 - 11:40 "Soil Features and Geologic Processes in the Alpine Tundra"- Mike Petersen and Tim Wheeler, Soil Scientists, Natural Resources Conservation Service Box Lunch 12:30 PM Depart 1:00 Arrive Many Parks Curve Interpretive Area (elevation 9,620 ft.) View of Valleys and Glacial Moraines, Photo Opportunity 1:30 Depart 3:00 Arrive Bobcat Gulch Fire Area, Arapaho-Roosevelt National Forest 3:10 "Fire History and Burned Area Emergency Rehabilitation Efforts" - Carl Chambers, U. S. Forest Service 3:40 "Involvement and Interaction With the Private Sector"- Todd Boldt; District Conservationist, Natural Resources Conservation Service 4:10 "Current Research on the Fire" - Colorado State University 4:45 Depart 6:00 Arrive Ft. Collins Marriott 2 3 Navigator’s Narrative Tim Wheeler Between the Fall River Visitors Center and the Lawn Lake Alluvial Debris Fan: This Park, or open grassy area, is called Horseshoe Park and is the tail end of the Park’s largest valley glacier. -

State of the Park Report
National Park Service U.S. Department of the Interior State of the Park Report Rocky Mountain National Park Colorado December 2017 National Park Service. 2017. State of the Park Report for Rocky Mountain National Park. State of the Park Series No. 50. National Park Service, Washington, DC. On the cover: Hallett Peak reflected in Dream Lake. NPS Photo. Disclaimer. This State of the Park report summarizes the current condition of park resources, visitor experience, and park infrastructure as assessed by a combination of available factual information and the expert opinion and professional judgment of park staff and subject matter experts. The internet version of this report provides additional details and sources of information about the findings summarized in the report, including references, accounts on the origin and quality of the data, and the methods and analytic approaches used in data collection and assessments of condition. This report provides evaluations of status and trends based on interpretation by NPS scientists and managers of both quantitative and non-quantitative assessments and observations. Future condition ratings may differ from findings in this report as new data and knowledge become available. The park superintendent approved the publication of this report. Executive Summary The mission of the National Park Service is to preserve unimpaired the natural and cultural resources and values of national parks for the enjoyment, education, and inspiration of this and future generations. NPS Management Policies (2006) state that “The Service will also strive to ensure that park resources and values are passed on to future generations in a condition that is as good as, or better than, the conditions that exist today.” As part of the stewardship of national parks for the American people, the NPS has begun to develop State of the Park reports to assess the overall status and trends of each park’s resources. -

Rocky Mountain National Park Trail System
Rocky Mountain National Park Trail Map HOURGLASS RESERVIOR Rocky M4ountain National Park Trail System 1 TRAP LAKE Y TWIN LAKE RESERVIOR W PETERSON LAKE H JOE WRIGHT RESERVIOR O L O C ZIMMERMAN LAKE MIRROR LAKE R E P P U , S S A P Y M Corral Creek USFS Trail Head M (! U M LAKE HUSTED 4 HWY 1 LOST LAKE COLO PPER LAKE LOUISE LOST LAKE, U #*Lost Falls Rowe Mountain LAKE DUNRAVEN LOST LAKE 13184 , LOWER Dunraven USFS Trail Head LONG DRAW RESERVIOR D (! Rowe Peak 13404 Hagues PeaDk 13560 D MICHIGAN LAKES TH LAKE AGNES E S SNOW LAKE La Poudre Pass Trail Head AD Mummy Mountain (! DL E 13425 D Fairchild Mountain 13502 D CRYSTAL LAKE LAWN LAKE TH UN Ypsilon Mountain DE R 13514 PA B SS D L A C R K PE C P SPECTACLE LAKES A , U N ER Chiquita, Mount Y IV D O R ST 13069 N E WE , DR IL U U A Y P O 4 TR P P P 3 TE Chapin Pass Trail Head S E Bridal Veil Falls LAKE OF THE CLOUDS Y U (! IL W O R #* H S N ER Cow Creek Trail Head U L K, LOW (! R A REE K OW C E C E V C(!rater Trail Head I (! U R POUDRE LAKE Cache La Poudre Trail Head S H O (! W D Milner Pass Trail Head Chasm Falls Y A #* R 3 Horseshoe Falls 4 Rock Cut Trail Head O ! #* L ( Thousand Falls O #* C Lawn Lake Trail Head FAN LAKE (! Colorado River Trail Head SHEEP LAKES (! Timber Lake Trail Head (! Beaver Ponds Trail Head (! CASCADE LAKE HIDDEN VALLEY BEAVER PONDS Lumpy Ridge Trail Head Ute Crossing Trail Head (! (! FOREST LAKE Deer Mountain/ Deer Ridge Trail Head ARROWHEAD LAKE ROCK LAKE (! U TE T TOWN OF RA LAKE ESTES IL Never Summer Trail Head INKWELL LAKE EA ESTES PARK (! ST U Upper Beaver Meadows -

A Guide to the Geology of Rocky Mountain National Park, Colorado
A Guide to the Geology of ROCKY MOUNTAIN NATIONAL PARK COLORADO For sale by the Superintendent of Documents, Washington, D. C. Price 15 cents A Guide to the Geology of ROCKY MOUNTAIN NATIONAL PARK [ COLORADO ] By Carroll H. Wegemann Former Regional Geologist, National Park Service UNITED STATES DEPARTMENT OF THE INTERIOR HAROLD L. ICKES, Secretary NATIONAL PARK SERVICE . NEWTON B. DRURY, Director UNITED STATES GOVERNMENT PRINTING OFFICE WASHINGTON : 1944 Table of Contents PAGE INTRODUCTION in BASIC FACTS ON GEOLOGY 1 THE OLDEST ROCKS OF THE PARK 2 THE FIRST MOUNTAINS 3 The Destruction of the First Mountains 3 NATURE OF PALEOZOIC DEPOSITS INDICATES PRESENCE OF SECOND MOUNTAINS 4 THE ROCKY MOUNTAINS 4 Time and Form of the Mountain Folding 5 Erosion Followed by Regional Uplift 5 Evidences of Intermittent Uplift 8 THE GREAT ICE AGE 10 Continental Glaciers 11 Valley Glaciers 11 POINTS OF INTEREST ALONG PARK ROADS 15 ROAD LOGS 18 Thompson River Entrance to Deer Ridge Junction 18 Deer Ridge Junction to Fall River Pass via Fall River .... 20 Fall River Pass to Poudre Lakes 23 Trail Ridge Road between Fall River Pass and Deer Ridge Junction 24 Deer Ridge Junction to Fall River Entrance via Horseshoe Park 29 Bear Lake Road 29 ILLUSTRATIONS LONGS PEAK FROM BEAR LAKE Front and back covers CHASM FALLS Inside back cover FIGURE PAGE 1. GEOLOGIC TIME SCALE iv 2. LONGS PEAK FROM THE EAST 3 3. PROFILE SECTION ACROSS THE ROCKY MOUNTAINS 5 4. ANCIENT EROSIONAL PLAIN ON TRAIL RIDGE 6 5. ANCIENT EROSIONAL PLAIN FROM FLATTOP MOUNTAIN ... 7 6. VIEW NORTHWEST FROM LONGS PEAK 8 7. -

Colorado Mountain Club 2009 Donors to CMC Annual Campaign [Oct
The Colorado Mountain Club ANNUAL REPORT 2009 Annual Report 2009 1 From the Chief Executive Officer he past year was an Dominguez Escalante National Conservation Area just south incredibly challenging time of Grand Junction. On the Front Range, we celebrated the for our entire country, designation of Rocky Mountain National Park’s backcountry Teconomically speaking. Businesses as wilderness. This designation is one of the final chapters in around the country failed, and the long journey of protecting the beloved park the CMC many people saw their personal played a major role in creating in 1915. finances change significantly. Every Our Youth Education Program introduced nearly once in awhile an event happens 5,000 youth and their chaperones to an education only the that makes all of us change our great outdoors can bring, and we furthered our work with focus and get back to basics. In the severely disabled children. I can’t tell you what an honor it nonprofit world, buckling down was for me to watch a young man who has been wheelchair while still achieving our goals is bound his entire life get to the top of our climbing wall. That not new to us; we always make experience alone gave me strength to get through last year’s magic happen with very little. I’m tough times. proud to report that the Colorado The upcoming year will be another time of growth Mountain Club saw a number of and change for the CMC. We are inching our way closer to achievements this past year despite our 100th anniversary and have begun a rebranding project the economic challenges. -

Rocky Mountain National Park High Country Headlines
Rocky Mountain National Park HIGH COUNTRY HEADLINES Winter 2006-07 October 29 - March 30 Your Park in Winter Reflected sunlight sparkles in the snow. Tracks of tiny mice and great elk cross INSIDE your trail. Frozen alpine lakes ringed 2 You Need to Know by massive peaks can be reached by snowshoe, ski, and even on foot. For 3 Survival those who are prepared, winter in Rocky Mountain National Park is a beautiful time 4 Ranger-led Programs full of crisp adventures. 5 Camping 6-7 Winter Tours 8 Park Map The park’s west side holds the best snow. photo: Harry Canon This newspaper is designed to help you most of the winter. Easy trails head toward drifts, Trail Ridge Road is too dangerous comfortably and safely enjoy this high Lulu City or Sun Valley, and many more to try to keep fully open through the and wild park during its longest season. challenging options are also available. On winter. Yet much of the park is still open Information on visitor centers, important the east side of the park (Estes Park area), year-round. You can drive to magnificent phone numbers, winter travel, and snowshoeing is more reliable than cross- view areas like Many Parks Curve and recreation are on pages 2 and 3. Free country skiing. The lofty peaks of Rocky Bear Lake on the east, and through the ranger-led programs are listed on page 4. Mountain National Park tend to catch and spectacular Kawuneeche Valley on the Camping is described on page 5. Some hold more snow on their western slopes west.