Qiu Et Al Table S1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Low-Temperature Effects on Docosahexaenoic Acid Biosynthesis
Hu et al. Biotechnol Biofuels (2020) 13:172 https://doi.org/10.1186/s13068-020-01811-y Biotechnology for Biofuels RESEARCH Open Access Low-temperature efects on docosahexaenoic acid biosynthesis in Schizochytrium sp. TIO01 and its proposed underlying mechanism Fan Hu1 , April L. Clevenger2, Peng Zheng3, Qiongye Huang1 and Zhaokai Wang1* Abstract Background: Schizochytrium species are known for their abundant production of docosahexaenoic acid (DHA). Low temperatures can promote the biosynthesis of polyunsaturated fatty acids (PUFAs) in many species. This study investi- gates low-temperature efects on DHA biosynthesis in Schizochytrium sp. TIO01 and its underlying mechanism. Results: The Schizochytrium fatty acid biosynthesis pathway was evaluated based on de novo genome assembly (contig N50 2.86 Mb) and iTRAQ-based protein identifcation. Our fndings revealed that desaturases, involved in DHA synthesis= via the fatty acid synthase (FAS) pathway, were completely absent. The polyketide synthase (PKS) path- way and the FAS pathway are, respectively, responsible for DHA and saturated fatty acid synthesis in Schizochytrium. Analysis of fatty acid composition profles indicates that low temperature has a signifcant impact on the production of DHA in Schizochytrium, increasing the DHA content from 43 to 65% of total fatty acids. However, the expression levels of PKS pathway genes were not signifcantly regulated as the DHA content increased. Further, gene expression analysis showed that pathways related to the production of substrates (acetyl-CoA and NADPH) for fatty acid synthe- sis (the branched-chain amino acid degradation pathway and the pentose phosphate pathway) and genes related to saturated fatty acid biosynthesis (the FAS pathway genes and malic enzyme) were, respectively, upregulated and downregulated. -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Test Catalogue August 2019
Test Catalogue August 2019 www.centogene.com/catalogue Table of Contents CENTOGENE CLINICAL DIAGNOSTIC PRODUCTS AND SERVICES › Whole Exome Testing 4 › Whole Genome Testing 5 › Genome wide CNV Analysis 5 › Somatic Mutation Analyses 5 › Biomarker Testing, Biochemical Testing 6 › Prenatal Testing 7 › Additional Services 7 › Metabolic Diseases 9 - 21 › Neurological Diseases 23 - 47 › Ophthalmological Diseases 49 - 55 › Ear, Nose and Throat Diseases 57 - 61 › Bone, Skin and Immune Diseases 63 - 73 › Cardiological Diseases 75 - 79 › Vascular Diseases 81 - 82 › Liver, Kidney and Endocrinological Diseases 83 - 89 › Reproductive Genetics 91 › Haematological Diseases 93 - 96 › Malformation and/or Retardation Syndromes 97 - 107 › Oncogenetics 109 - 113 ® › CentoXome - Sequencing targeting exonic regions of ~20.000 genes Test Test name Description code CentoXome® Solo Medical interpretation/report of WES findings for index 50029 CentoXome® Solo - Variants Raw data; fastQ, BAM, Vcf files along with variant annotated file in xls format for index 50028 CentoXome® Solo - with CNV Medical interpretation/report of WES including CNV findings for index 50103 Medical interpretation/report of WES in index, package including genome wide analyses of structural/ CentoXome® Solo - with sWGS 50104 large CNVs through sWGS Medical interpretation/report of WES in index, package including genome wide analyses of structural/ CentoXome® Solo - with aCGH 750k 50122 large CNVs through 750k microarray Medical interpretation/report of WES in index, package including genome -

Proteomic Characterisation of Rat Bone Marrow-Derived Mesenchymal Stromal Cells Cultured in ‘Stemness’ Promoting Conditions
PROTEOMIC CHARACTERISATION OF RAT BONE MARROW-DERIVED MESENCHYMAL STROMAL CELLS CULTURED IN ‘STEMNESS’ PROMOTING CONDITIONS. Morgan Madeline Carlton Bachelor of Biomedical Science, QUT 2015 Tissue Repair and Translational Physiology Program Institute of Health and Biomedical Innovation School of Biomedical Sciences Faculty of Health Queensland University of Technology Submitted in fulfilment of the requirements for the degree of Master of Philosophy (Health) 2018 Keywords Biological processes Bone marrow-derived mesenchymal stromal cells Bone morphogenetic protein 4 Data independent acquisition Differentiation Fibroblast growth factor 2 Fibronectin Foetal calf serum Gene Ontology Mass spectrometry Mesenchymal stromal cells MetaboAnalyst OFFGEL electrophoresis Protein profiling Proteomics Rat Serum concentration Stemness SWATH acquisition Proteomic Characterisation of Rat Bone Marrow-Derived Mesenchymal Stromal Cells Cultured in ‘Stemness’ Promoting Conditions. i Abstract Mesenchymal stromal cells (MSCs) have the potential to be powerful medical tools for utilisation in clinical practice, for the treatment of numerous ailments including wound healing, tissue regeneration and potentially even spinal injury repair. To maximise the use of MSCs, access to an off-the-shelf MSC therapeutic is highly desirable, however, achieving this outcome is challenging, as MSCs cannot survive, undifferentiated in culture for extended periods of time. Currently, research in this field aims to determine an optimal culture environment for the growth of MSCs, that maintains their stromal phenotype. Evaluation of MSC differentiation and stemness have been investigated using methods of proteomics previously however, advanced protein profiling methods, such as SWATH-MS, have yet to be utilised for this purpose. Therefore, the purpose of this study was to investigate the effect that specific growth factors had on MSCs during culture through the utilisation of SWATH-MS analysis, with specific interest in the maintenance of their undifferentiated state. -

Download CGT Exome V2.0
CGT Exome version 2. -

Proteomic Analysis of Aged Microglia
Flowers et al. Journal of Neuroinflammation (2017) 14:96 DOI 10.1186/s12974-017-0840-7 RESEARCH Open Access Proteomic analysis of aged microglia: shifts in transcription, bioenergetics, and nutrient response Antwoine Flowers1,2, Harris Bell-Temin3, Ahmad Jalloh1,2, Stanley M. Stevens Jr.3 and Paula C. Bickford1,2,4* Abstract Background: Age is the primary risk factor for many diseases. As such, age is a critical co-factor for examination in order to understand the progression and potential intervention in disease progression. Studies examining both the phenotype and transcriptome of aged microglia demonstrated a propensity for the development of a pro-inflammatory phenotype. Less well studied is the concomitant blunting of anti-inflammatory aspects of microglial function with age which also impact plasticity and repair in the CNS. Methods: This study utilizes mass spectrometry-based proteomics to compare primary microglia from young andagedanimals. Results: This study revealed alterations in three clusters of inter-related proteins. The three pathways were inflammatory signaling, mitochondrial function, and cellular metabolism. Analysis of these clusters identified the protein rapamycin-insensitive companion of mTOR (RICTOR), a component of the mTORC2 complex, as a novel upstream regulator of several biological functions that are altered with age and potentially linked to phenotype development. A decrease in mTORC2-dependent AKT S473 phosphorylation, as assessed by insulin growth factor (IGF) treatment, was observed in aged microglia. This novel finding was confirmed by genetic manipulation of the microglial cell line. BV2 cells with diminished RICTOR displayed a phenotype that was strikingly similar to that of aged microglia. This finding is particularly relevant as the mTOR pathway already has a number of pharmacological modulators used clinically. -

Spatial Sorting Enables Comprehensive Characterization of Liver Zonation
ARTICLES https://doi.org/10.1038/s42255-019-0109-9 Spatial sorting enables comprehensive characterization of liver zonation Shani Ben-Moshe1,3, Yonatan Shapira1,3, Andreas E. Moor 1,2, Rita Manco1, Tamar Veg1, Keren Bahar Halpern1 and Shalev Itzkovitz 1* The mammalian liver is composed of repeating hexagonal units termed lobules. Spatially resolved single-cell transcriptomics has revealed that about half of hepatocyte genes are differentially expressed across the lobule, yet technical limitations have impeded reconstructing similar global spatial maps of other hepatocyte features. Here, we show how zonated surface markers can be used to sort hepatocytes from defined lobule zones with high spatial resolution. We apply transcriptomics, microRNA (miRNA) array measurements and mass spectrometry proteomics to reconstruct spatial atlases of multiple zon- ated features. We demonstrate that protein zonation largely overlaps with messenger RNA zonation, with the periportal HNF4α as an exception. We identify zonation of miRNAs, such as miR-122, and inverse zonation of miRNAs and their hepa- tocyte target genes, highlighting potential regulation of gene expression levels through zonated mRNA degradation. Among the targets, we find the pericentral Wingless-related integration site (Wnt) receptors Fzd7 and Fzd8 and the periportal Wnt inhibitors Tcf7l1 and Ctnnbip1. Our approach facilitates reconstructing spatial atlases of multiple cellular features in the liver and other structured tissues. he mammalian liver is a structured organ, consisting of measurements would broaden our understanding of the regulation repeating hexagonally shaped units termed ‘lobules’ (Fig. 1a). of liver zonation and could be used to model liver metabolic func- In mice, each lobule consists of around 9–12 concentric lay- tion more precisely. -

Loss of the E3 Ubiquitin Ligase MKRN1 Represses Diet-Induced Metabolic Syndrome Through AMPK Activation
ARTICLE DOI: 10.1038/s41467-018-05721-4 OPEN Loss of the E3 ubiquitin ligase MKRN1 represses diet-induced metabolic syndrome through AMPK activation Min-Sik Lee1, Hyun-Ji Han2, Su Yeon Han2, Il Young Kim3,4, Sehyun Chae5, Choong-Sil Lee2, Sung Eun Kim2, Seul Gi Yoon4, Jun-Won Park4, Jung-Hoon Kim2, Soyeon Shin2, Manhyung Jeong2, Aram Ko2, Ho-Young Lee6, Kyoung-Jin Oh 7, Yun-Hee Lee 8, Kwang-Hee Bae7, Seung-Hoi Koo9, Jea-woo Kim10, Je Kyung Seong3,4, Daehee Hwang5 & Jaewhan Song 2 1234567890():,; AMP-activated protein kinase (AMPK) plays a key role in controlling energy metabolism in response to physiological and nutritional status. Although AMPK activation has been pro- posed as a promising molecular target for treating obesity and its related comorbidities, the use of pharmacological AMPK activators has been met with contradictory therapeutic challenges. Here we show a regulatory mechanism for AMPK through its ubiquitination and degradation by the E3 ubiquitin ligase makorin ring finger protein 1 (MKRN1). MKRN1 depletion promotes glucose consumption and suppresses lipid accumulation due to AMPK stabilisation and activation. Accordingly, MKRN1-null mice show chronic AMPK activation in both liver and adipose tissue, resulting in significant suppression of diet-induced metabolic syndrome. We demonstrate also its therapeutic effect by administering shRNA targeting MKRN1 into obese mice that reverses non-alcoholic fatty liver disease. We suggest that ubiquitin-dependent AMPK degradation represents a target therapeutic strategy for meta- bolic disorders. 1 Harvard Medical School, Boston Children’s Hospital, 3 Blackfan Circle CLS-16060.2, Boston, MA 02115, USA. 2 Department of Biochemistry, College of Life Science and Biotechnology, Yonsei University, Seoul 03722, Republic of Korea. -

Proteomic Anaysis of Aged Microglia: Shifts in Transcription, Bioenergetics, and Nutrient Response Antwoine Flowers1,2, Harris Bell-Temin3, Ahmad Jalloh1,2, Stanley M
Flowers et al. Journal of Neuroinflammation (2017) 14:96 DOI 10.1186/s12974-017-0840-7 RESEARCH Open Access Proteomic anaysis of aged microglia: shifts in transcription, bioenergetics, and nutrient response Antwoine Flowers1,2, Harris Bell-Temin3, Ahmad Jalloh1,2, Stanley M. Stevens Jr.3 and Paula C. Bickford1,2,4* Abstract Background: Age is the primary risk factor for many diseases. As such, age is a critical co-factor for examination in order to understand the progression and potential intervention in disease progression. Studies examining both the phenotype and transcriptome of aged microglia demonstrated a propensity for the development of a pro-inflammatory phenotype. Less well studied is the concomitant blunting of anti-inflammatory aspects of microglial function with age which also impact plasticity and repair in the CNS. Methods: This study utilizes mass spectrometry-based proteomics to compare primary microglia from young andagedanimals. Results: This study revealed alterations in three clusters of inter-related proteins. The three pathways were inflammatory signaling, mitochondrial function, and cellular metabolism. Analysis of these clusters identified the protein rapamycin-insensitive companion of mTOR (RICTOR), a component of the mTORC2 complex, as a novel upstream regulator of several biological functions that are altered with age and potentially linked to phenotype development. A decrease in mTORC2-dependent AKT S473 phosphorylation, as assessed by insulin growth factor (IGF) treatment, was observed in aged microglia. This novel finding was confirmed by genetic manipulation of the microglial cell line. BV2 cells with diminished RICTOR displayed a phenotype that was strikingly similar to that of aged microglia. This finding is particularly relevant as the mTOR pathway already has a number of pharmacological modulators used clinically. -
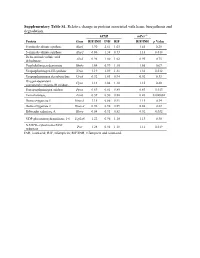
Supplementary Table S1. Relative Change in Proteins Associated with Heme Biosynthesis and Degradation
Supplementary Table S1. Relative change in proteins associated with heme biosynthesis and degradation. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value 5-aminolevulinate synthase Alas1 1.90 2.61 1.05 1.41 0.28 5-aminolevulinate synthase Alas2 0.86 1.38 0.73 1.18 0.018 Delta-aminolevulinic acid Alad 0.96 1.00 1.02 0.95 0.75 dehydratase Porphobilinogen deaminase Hmbs 1.04 0.99 1.10 1.05 0.67 Uroporphyrinogen-III synthase Uros 1.19 1.09 1.31 1.38 0.012 Uroporphyrinogen decarboxylase Urod 0.92 1.03 0.94 0.92 0.33 Oxygen-dependent Cpox 1.13 1.04 1.18 1.15 0.20 coproporphyrinogen-III oxidase, Protoporphyrinogen oxidase Ppox 0.69 0.81 0.85 0.83 0.013 Ferrochelatase, Fech 0.39 0.50 0.88 0.43 0.000002 Heme oxygenase 1 Hmox1 1.15 0.86 0.91 1.11 0.34 Heme oxygenase 2 Hmox2 0.96 0.98 0.89 0.88 0.22 Biliverdin reductase A Blvra 0.84 0.92 0.82 0.92 0.032 UDP-glucuronosyltransferase 1-6 Ugt1a6 1.22 0.96 1.10 1.13 0.30 NADPH--cytochrome P450 Por 1.28 0.92 1.18 1.12 0.019 reductase INH, isoniazid; RIF, rifampicin; RIF/INH, rifampicin and isoniazid. Supplementary Table S2. Relative change in protein nuclear receptors. hPXR mPxr–/– Protein Gene RIF/INH INH RIF RIF/INH p Value Aryl hydrocarbon receptor Ahr 1.09 0.91 1.00 1.26 0.092 Hepatocyte nuclear factor Hnf1a 0.87 0.97 0.82 0.79 0.027 1-alpha Hepatocyte nuclear factor Hnf4a 0.95 1.05 0.97 1.08 0.20 4-alpha Oxysterols receptor LXR- Nr1h3 0.94 1.16 1.03 1.02 0.42 alpha Bile acid receptor Nr1h4 1.05 1.17 0.98 1.19 0.12 Retinoic acid receptor Rxra 0.88 1.03 0.83 0.95 0.12 RXR-alpha Peroxisome proliferator-