Thesis.Pdf (5.857Mb)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Uwsgi Documentation Release 1.9
uWSGI Documentation Release 1.9 uWSGI February 08, 2016 Contents 1 Included components (updated to latest stable release)3 2 Quickstarts 5 3 Table of Contents 11 4 Tutorials 137 5 Articles 139 6 uWSGI Subsystems 141 7 Scaling with uWSGI 197 8 Securing uWSGI 217 9 Keeping an eye on your apps 223 10 Async and loop engines 231 11 Web Server support 237 12 Language support 251 13 Release Notes 317 14 Contact 359 15 Donate 361 16 Indices and tables 363 Python Module Index 365 i ii uWSGI Documentation, Release 1.9 The uWSGI project aims at developing a full stack for building (and hosting) clustered/distributed network applica- tions. Mainly targeted at the web and its standards, it has been successfully used in a lot of different contexts. Thanks to its pluggable architecture it can be extended without limits to support more platforms and languages. Cur- rently, you can write plugins in C, C++ and Objective-C. The “WSGI” part in the name is a tribute to the namesake Python standard, as it has been the first developed plugin for the project. Versatility, performance, low-resource usage and reliability are the strengths of the project (and the only rules fol- lowed). Contents 1 uWSGI Documentation, Release 1.9 2 Contents CHAPTER 1 Included components (updated to latest stable release) The Core (implements configuration, processes management, sockets creation, monitoring, logging, shared memory areas, ipc, cluster membership and the uWSGI Subscription Server) Request plugins (implement application server interfaces for various languages and platforms: WSGI, PSGI, Rack, Lua WSAPI, CGI, PHP, Go ...) Gateways (implement load balancers, proxies and routers) The Emperor (implements massive instances management and monitoring) Loop engines (implement concurrency, components can be run in preforking, threaded, asynchronous/evented and green thread/coroutine modes. -

NGINX-Conf-2018-Slides Rawdat
Performance Tuning NGINX Name: Amir Rawdat Currently: Technical Marketing Engineer at NGINX inc. Previously: - Customer Applications Engineer at Nokia inc. Multi-Process Architecture with QPI Bus Web Server Topology wrk nginx Reverse Proxy Topology wrk nginx nginx J6 Technical Specifications # Sockets # Cores # Model RAM OS NIC per Threads Name Socket per Core Client 2 22 2 Intel(R) 128 GB Ubuntu 40GbE Xeon(R) CPU Xenial QSFP+ E5-2699 v4 @ 2.20GHz Web Server 2 24 2 Intel(R) 192 GB Ubuntu 40GbE Xeon(R) & Platinum Xenial QSFP+ Reverse 8168 CPU @ Proxy 2.70GHz Multi-Processor Architecture #1 Duplicate NGINX Configurations J9 Multi-Processor Architecture NGINX Configuration (Instance 1) user root; worker_processes 48 ; worker_cpu_affinity auto 000000000000000000000000111111111111111111111111000000000000000000000000111111111111111111111111; worker_rlimit_nofile 1024000; error_log /home/ubuntu/access.error error; ….. ……. J11 NGINX Configuration (Instance 2) user root; worker_processes 48 ; worker_cpu_affinity auto 111111111111111111111111000000000000000000000000111111111111111111111111000000000000000000000000; worker_rlimit_nofile 1024000; error_log /home/ubuntu/access.error error; ……. ……. J12 Deploying NGINX Instances $ nginx –c /path/to/configuration/instance-1 $ nginx –c /path/to/configuration/instance-2 $ ps aux | grep nginx nginx: master process /usr/sbin/nginx -c /etc/nginx/nginx_0.conf nginx: worker process nginx: worker process nginx: master process /usr/sbin/nginx -c /etc/nginx/nginx_1.conf nginx: worker process nginx: worker process -

Bepasty Documentation Release 0.3.0
bepasty Documentation Release 0.3.0 The Bepasty Team (see AUTHORS file) Jul 02, 2019 Contents 1 Contents 3 1.1 bepasty..................................................3 1.2 Using bepasty’s web interface......................................4 1.3 Using bepasty with non-web clients...................................6 1.4 Quickstart................................................7 1.5 Installation tutorial with Debian, NGinx and gunicorn......................... 10 1.6 ChangeLog................................................ 12 1.7 The bepasty software Project....................................... 14 1.8 License.................................................. 14 1.9 Authors.................................................. 15 Index 17 i ii bepasty Documentation, Release 0.3.0 bepasty is like a pastebin for every kind of file (text, image, audio, video, documents, . ). You can upload multiple files at once, simply by drag and drop. Contents 1 bepasty Documentation, Release 0.3.0 2 Contents CHAPTER 1 Contents 1.1 bepasty bepasty is like a pastebin for all kinds of files (text, image, audio, video, documents, . , binary). The documentation is there: http://bepasty-server.readthedocs.org/en/latest/ 1.1.1 Features • Generic: – you can upload multiple files at once, simply by drag and drop – after upload, you get a unique link to a view of each file – on that view, we show actions you can do with the file, metadata of the file and, if possible, we also render the file contents – if you uploaded multiple files, you can create a pastebin with the list -

Next Generation Web Scanning Presentation
Next generation web scanning New Zealand: A case study First presented at KIWICON III 2009 By Andrew Horton aka urbanadventurer NZ Web Recon Goal: To scan all of New Zealand's web-space to see what's there. Requirements: – Targets – Scanning – Analysis Sounds easy, right? urbanadventurer (Andrew Horton) www.morningstarsecurity.com Targets urbanadventurer (Andrew Horton) www.morningstarsecurity.com Targets What does 'NZ web-space' mean? It could mean: •Geographically within NZ regardless of the TLD •The .nz TLD hosted anywhere •All of the above For this scan it means, IPs geographically within NZ urbanadventurer (Andrew Horton) www.morningstarsecurity.com Finding Targets We need creative methods to find targets urbanadventurer (Andrew Horton) www.morningstarsecurity.com DNS Zone Transfer urbanadventurer (Andrew Horton) www.morningstarsecurity.com Find IP addresses on IRC and by resolving lots of NZ websites 58.*.*.* 60.*.*.* 65.*.*.* 91.*.*.* 110.*.*.* 111.*.*.* 113.*.*.* 114.*.*.* 115.*.*.* 116.*.*.* 117.*.*.* 118.*.*.* 119.*.*.* 120.*.*.* 121.*.*.* 122.*.*.* 123.*.*.* 124.*.*.* 125.*.*.* 130.*.*.* 131.*.*.* 132.*.*.* 138.*.*.* 139.*.*.* 143.*.*.* 144.*.*.* 146.*.*.* 150.*.*.* 153.*.*.* 156.*.*.* 161.*.*.* 162.*.*.* 163.*.*.* 165.*.*.* 166.*.*.* 167.*.*.* 192.*.*.* 198.*.*.* 202.*.*.* 203.*.*.* 210.*.*.* 218.*.*.* 219.*.*.* 222.*.*.* 729,580,500 IPs. More than we want to try. urbanadventurer (Andrew Horton) www.morningstarsecurity.com IP address blocks in the IANA IPv4 Address Space Registry Prefix Designation Date Whois Status [1] ----- -

Load Balancing for Heterogeneous Web Servers
Load Balancing for Heterogeneous Web Servers Adam Pi´orkowski1, Aleksander Kempny2, Adrian Hajduk1, and Jacek Strzelczyk1 1 Department of Geoinfomatics and Applied Computer Science, AGH University of Science and Technology, Cracow, Poland {adam.piorkowski,jacek.strzelczyk}@agh.edu.pl http://www.agh.edu.pl 2 Adult Congenital and Valvular Heart Disease Center University of Muenster, Muenster, Germany [email protected] http://www.ukmuenster.de Abstract. A load balancing issue for heterogeneous web servers is de- scribed in this article. The review of algorithms and solutions is shown. The selected Internet service for on-line echocardiography training is presented. The independence of simultaneous requests for this server is proved. Results of experimental tests are presented3. Key words: load balancing, scalability, web server, minimum response time, throughput, on-line simulator 1 Introduction Modern web servers can handle millions of queries, although the performance of a single node is limited. Performance can be continuously increased, if the services are designed so that they can be scaled. The concept of scalability is closely related to load balancing. This technique has been used since the beginning of the first distributed systems, including rich client architecture. Most of the complex web systems use load balancing to improve performance, availability and security [1{4]. 2 Load Balancing in Cluster of web servers Clustering of web servers is a method of constructing scalable Internet services. The basic idea behind the construction of such a service is to set the relay server 3 This is the accepted version of: Piorkowski, A., Kempny, A., Hajduk, A., Strzelczyk, J.: Load Balancing for Heterogeneous Web Servers. -

Zope Documentation Release 5.3
Zope Documentation Release 5.3 The Zope developer community Jul 31, 2021 Contents 1 What’s new in Zope 3 1.1 What’s new in Zope 5..........................................4 1.2 What’s new in Zope 4..........................................4 2 Installing Zope 11 2.1 Prerequisites............................................... 11 2.2 Installing Zope with zc.buildout .................................. 12 2.3 Installing Zope with pip ........................................ 13 2.4 Building the documentation with Sphinx ............................... 14 3 Configuring and Running Zope 15 3.1 Creating a Zope instance......................................... 16 3.2 Filesystem Permissions......................................... 17 3.3 Configuring Zope............................................. 17 3.4 Running Zope.............................................. 18 3.5 Running Zope (plone.recipe.zope2instance install)........................... 20 3.6 Logging In To Zope........................................... 21 3.7 Special access user accounts....................................... 22 3.8 Troubleshooting............................................. 22 3.9 Using alternative WSGI server software................................. 22 3.10 Debugging Zope applications under WSGI............................... 26 3.11 Zope configuration reference....................................... 27 4 Migrating between Zope versions 37 4.1 From Zope 2 to Zope 4 or 5....................................... 37 4.2 Migration from Zope 4 to Zope 5.0.................................. -

Generator of Slow Denial-Of-Service Cyber Attacks †
sensors Article Generator of Slow Denial-of-Service Cyber Attacks † Marek Sikora * , Radek Fujdiak , Karel Kuchar , Eva Holasova and Jiri Misurec Department of Telecommunications, Faculty of Electrical Engineering and Communications, Brno University of Technology, Technicka 12, 616 00 Brno, Czech Republic; [email protected] (R.F.); [email protected] (K.K.); [email protected] (E.H.); [email protected] (J.M.) * Correspondence: [email protected] † This paper is an extended version of our paper published in International Congress on Ultra Modern Telecommunications and Control Systems (ICUMT 2020), Brno, Czech Republic, 5–7 October 2020. Abstract: In today’s world, the volume of cyber attacks grows every year. These attacks can cause many people or companies high financial losses or loss of private data. One of the most common types of attack on the Internet is a DoS (denial-of-service) attack, which, despite its simplicity, can cause catastrophic consequences. A slow DoS attack attempts to make the Internet service unavailable to users. Due to the small data flows, these attacks are very similar to legitimate users with a slow Internet connection. Accurate detection of these attacks is one of the biggest challenges in cybersecurity. In this paper, we implemented our proposal of eleven major and most dangerous slow DoS attacks and introduced an advanced attack generator for testing vulnerabilities of protocols, servers, and services. The main motivation for this research was the absence of a similarly comprehensive generator for testing slow DoS vulnerabilities in network systems. We built an experimental environment for testing our generator, and then we performed a security analysis of the five most used web servers. -

Uwsgi Documentation Release 2.0
uWSGI Documentation Release 2.0 uWSGI Jun 17, 2020 Contents 1 Included components (updated to latest stable release)3 2 Quickstarts 5 3 Table of Contents 33 4 Tutorials 303 5 Articles 343 6 uWSGI Subsystems 375 7 Scaling with uWSGI 457 8 Securing uWSGI 485 9 Keeping an eye on your apps 503 10 Async and loop engines 511 11 Web Server support 525 12 Language support 541 13 Other plugins 629 14 Broken/deprecated features 633 15 Release Notes 643 16 Contact 741 17 Commercial support 743 18 Donate 745 19 Sponsors 747 20 Indices and tables 749 i Python Module Index 751 Index 753 ii uWSGI Documentation, Release 2.0 The uWSGI project aims at developing a full stack for building hosting services. Application servers (for various programming languages and protocols), proxies, process managers and monitors are all implemented using a common api and a common configuration style. Thanks to its pluggable architecture it can be extended to support more platforms and languages. Currently, you can write plugins in C, C++ and Objective-C. The “WSGI” part in the name is a tribute to the namesake Python standard, as it has been the first developed plugin for the project. Versatility, performance, low-resource usage and reliability are the strengths of the project (and the only rules fol- lowed). Contents 1 uWSGI Documentation, Release 2.0 2 Contents CHAPTER 1 Included components (updated to latest stable release) The Core (implements configuration, processes management, sockets creation, monitoring, logging, shared memory areas, ipc, cluster membership and the uWSGI Subscription Server) Request plugins (implement application server interfaces for various languages and platforms: WSGI, PSGI, Rack, Lua WSAPI, CGI, PHP, Go . -

Introduction
HTTP Request Smuggling in 2020 – New Variants, New Defenses and New Challenges Amit Klein SafeBreach Labs Introduction HTTP Request Smuggling (AKA HTTP Desyncing) is an attack technique that exploits different interpretations of a stream of non-standard HTTP requests among various HTTP devices between the client (attacker) and the server (including the server itself). Specifically, the attacker manipulates the way various HTTP devices split the stream into individual HTTP requests. By doing this, the attacker can “smuggle” a malicious HTTP request through an HTTP device to the server abusing the discrepancy in the interpretation of the stream of requests and desyncing between the server’s view of the HTTP request (and response) stream and the intermediary HTTP device’s view of these streams. In this way, for example, the malicious HTTP request can be "smuggled" as a part of the previous HTTP request. HTTP Request Smuggling was invented in 2005, and recently, additional research cropped up. This research field is still not fully explored, especially when considering open source defense systems such as mod_security’s community rule-set (CRS). These HTTP Request Smuggling defenses are rudimentary and not always effective. My Contribution My contribution is three-fold. I explore new attacks and defense mechanisms, and I provide some “challenges”. 1. New attacks: I provide some new HTTP Request Smuggling variants and show how they work against various proxy-server (or proxy-proxy) combinations. I also found a bypass for mod_security CRS (assuming HTTP Request Smuggling is possible without it). An attack demonstration script implementing my payloads is available in SafeBreach Labs’ GitHub repository (https://github.com/SafeBreach-Labs/HRS). -

Owncloud Administrators Manual Release 7.0
ownCloud Administrators Manual Release 7.0 The ownCloud developers November 24, 2014 CONTENTS 1 Introduction 1 1.1 Target Audience.............................................1 1.2 Document Structure...........................................1 2 What’s New for Admins in ownCloud 75 2.1 New User Management.........................................5 2.2 External Storage.............................................5 2.3 Object Stores as Primary Storage....................................5 2.4 Server to Server Sharing.........................................5 2.5 SharePoint Integration (Enterprise Edition only)............................5 2.6 Windows Network Drive Integration (Enterprise Edition only).....................6 2.7 Sharing..................................................6 2.8 Email Configuration Wizard.......................................6 2.9 Editable Email Templates........................................6 2.10 Active Directory and LDAP Enhancements...............................7 3 Installation 9 3.1 ownCloud Appliances..........................................9 3.2 Installing and Managing Apps......................................9 3.3 Hiawatha Configuration......................................... 12 3.4 Installation Wizard............................................ 12 3.5 Lighttpd Configuration.......................................... 16 3.6 Linux Distributions............................................ 16 3.7 Mac OS X................................................ 17 3.8 Nginx Configuration.......................................... -
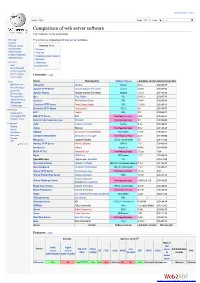
Comparison of Web Server Software from Wikipedia, the Free Encyclopedia
Create account Log in Article Talk Read Edit ViewM ohrisetory Search Comparison of web server software From Wikipedia, the free encyclopedia Main page This article is a comparison of web server software. Contents Featured content Contents [hide] Current events 1 Overview Random article 2 Features Donate to Wikipedia 3 Operating system support Wikimedia Shop 4 See also Interaction 5 References Help 6 External links About Wikipedia Community portal Recent changes Overview [edit] Contact page Tools Server Developed by Software license Last stable version Latest release date What links here AOLserver NaviSoft Mozilla 4.5.2 2012-09-19 Related changes Apache HTTP Server Apache Software Foundation Apache 2.4.10 2014-07-21 Upload file Special pages Apache Tomcat Apache Software Foundation Apache 7.0.53 2014-03-30 Permanent link Boa Paul Phillips GPL 0.94.13 2002-07-30 Page information Caudium The Caudium Group GPL 1.4.18 2012-02-24 Wikidata item Cite this page Cherokee HTTP Server Álvaro López Ortega GPL 1.2.103 2013-04-21 Hiawatha HTTP Server Hugo Leisink GPLv2 9.6 2014-06-01 Print/export Create a book HFS Rejetto GPL 2.2f 2009-02-17 Download as PDF IBM HTTP Server IBM Non-free proprietary 8.5.5 2013-06-14 Printable version Internet Information Services Microsoft Non-free proprietary 8.5 2013-09-09 Languages Jetty Eclipse Foundation Apache 9.1.4 2014-04-01 Čeština Jexus Bing Liu Non-free proprietary 5.5.2 2014-04-27 Galego Nederlands lighttpd Jan Kneschke (Incremental) BSD variant 1.4.35 2014-03-12 Português LiteSpeed Web Server LiteSpeed Technologies Non-free proprietary 4.2.3 2013-05-22 Русский Mongoose Cesanta Software GPLv2 / commercial 5.5 2014-10-28 中文 Edit links Monkey HTTP Server Monkey Software LGPLv2 1.5.1 2014-06-10 NaviServer Various Mozilla 1.1 4.99.6 2014-06-29 NCSA HTTPd Robert McCool Non-free proprietary 1.5.2a 1996 Nginx NGINX, Inc. -

Go for Sres Using Python
Go for SREs Using Python Andrew Hamilton What makes Go fun to work with? Go is expressive, concise, clean, and efficient It's a fast, statically typed, compiled language that feels like a dynamically typed, interpreted language Relatively new but stable Easy to build CLI tool or API quickly Fast garbage collection https://golang.org/doc/ Hello world // Go #!/usr/bin/env python3 package main # python3 import “fmt” def main(): print(“Hello world”) func main() { fmt.Println(“Hello world”) if __name__ == “__main__”: } main() $ go build hello_world.go $ python3 hello_world.py $ ./hello_world Hello world Hello world $ $ Good default tool chain Formatting (gofmt and go import) [pylint] Testing (go test) [pytest, nose] Race Detection (go build -race) Source Code Checking (go vet and go oracle) Help with refactoring (go refactor) BUT... There’s currently no official debugger Debuggers can be very helpful There are some unofficial debuggers https://github.com/mailgun/godebug Standard library Does most of what you’ll need already Built for concurrency and parallel processing where useful Quick to add and update features (HTTP2 support) but doesn’t break your code Expanding third-party libraries Third-party libraries are just other repositories Easy to build and propagate by pushing to Github or Bitbucket, etc List of dependencies not kept in a separate file but pulled from imports Versioning dependencies is done by vendoring within your project repository No PyPI equivalent Importing import ( “fmt” “net/http” “github.com/ahamilton55/some_lib/helpers”