University of Sheffield Department of Chemical and Biological Engineering
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DNA Topoisomerases and Cancer Topoisomerases and TOP Genes in Humans Humans Vs
DNA Topoisomerases Review DNA Topoisomerases And Cancer Topoisomerases and TOP Genes in Humans Humans vs. Escherichia Coli Topoisomerase differences Comparisons Topoisomerase and genomes Top 1 Top1 and Top2 differences Relaxation of DNA Top1 DNA supercoiling DNA supercoiling In the context of chromatin, where the rotation of DNA is constrained, DNA supercoiling (over- and under-twisting and writhe) is readily generated. TOP1 and TOP1mt remove supercoiling by DNA untwisting, acting as “swivelases”, whereas TOP2a and TOP2b remove writhe, acting as “writhases” at DNA crossovers (see TOP2 section). Here are some basic facts concerning DNA supercoiling that are relevant to topoisomerase activity: • Positive supercoiling (Sc+) tightens the DNA helix whereas negative supercoiling (Sc-) facilitates the opening of the duplex and the generation of single-stranded segments. • Nucleosome formation and disassembly absorbs and releases Sc-, respectively. • Polymerases generate Sc+ ahead and Sc- behind their tracks. • Excess of Sc+ arrests DNA tracking enzymes (helicases and polymerases), suppresses transcription elongation and initiation, and destabilizes nucleosomes. • Sc- facilitates DNA melting during the initiation of replication and transcription, D-loop formation and homologous recombination and nucleosome formation. • Excess of Sc- favors the formation of alternative DNA structures (R-loops, guanine quadruplexes, right-handed DNA (Z-DNA), plectonemic structures), which then absorb Sc- upon their formation and attract regulatory proteins. The -

Comparative Biochemistry and Physiology, Part D, Vol. 5, Pp. 45-54 (2010)
Comparative genomics and proteomics of vertebrate diacylglycerol acyltransferase (DGAT), acyl CoA wax alcohol acyltransferase (AWAT) and monoacylglycerol acyltransferase (MGAT) Author Holmes, Roger S Published 2010 Journal Title Comparative Biochemistry and Physiology, Part D DOI https://doi.org/10.1016/j.cbd.2009.09.004 Copyright Statement © 2010 Elsevier. This is the author-manuscript version of this paper. Reproduced in accordance with the copyright policy of the publisher. Please refer to the journal's website for access to the definitive, published version. Downloaded from http://hdl.handle.net/10072/36786 Griffith Research Online https://research-repository.griffith.edu.au Comparative Biochemistry and Physiology, Part D, Vol. 5, pp. 45-54 (2010) COMPARATIVE GENOMICS AND PROTEOMICS OF VERTEBRATE DIACYLGLYCEROL ACYLTRANSFERASE (DGAT), ACYL CoA WAX ALCOHOL ACYLTRANSFERASE (AWAT) AND MONOACYLGLYCEROL ACYLTRANSFERASE (MGAT) Roger S Holmes School of Biomolecular and Physical Sciences, Griffith University, Nathan 4111 Brisbane Queensland Australia Email: [email protected] Keywords: Diacylglycerol acyltransferase-Monoacylglycerol transferase-Human- Mouse-Opossum-Zebrafish-Genetics-Evolution-X chromosome Running Head: Genomics and proteomics of vertebrate acylglycerol acyltransferases ABSTRACT BLAT (BLAST-Like Alignment Tool) analyses of the opossum (Monodelphis domestica) and zebrafish (Danio rerio) genomes were undertaken using amino acid sequences of the acylglycerol acyltransferase (AGAT) superfamily. Evidence is reported for 8 opossum monoacylglycerol acyltransferase-like (MGAT) (E.C. 2.3.1.22) and diacylglycerol acyltransferase-like (DGAT) (E.C. 2.3.1.20) genes and proteins, including DGAT1, DGAT2, DGAT2L6 (DGAT2-like protein 6), AWAT1 (acyl-CoA wax alcohol acyltransferase 1), AWAT2, MGAT1, MGAT2 and MGAT3. Three of these genes (AWAT1, AWAT2 and DGAT2L6) are closely localized on the opossum X chromosome. -

Type IA Topoisomerases As Targets for Infectious Disease Treatments
microorganisms Review Type IA Topoisomerases as Targets for Infectious Disease Treatments Ahmed Seddek 1,2 , Thirunavukkarasu Annamalai 1 and Yuk-Ching Tse-Dinh 1,2,* 1 Department of Chemistry and Biochemistry, Florida International University, Miami, FL 33199, USA; asedd001@fiu.edu (A.S.); athiruna@fiu.edu (T.A.) 2 Biomolecular Sciences Institute, Florida International University, Miami, FL 33199, USA * Correspondence: ytsedinh@fiu.edu; Tel.: +1-305-348-4956 Abstract: Infectious diseases are one of the main causes of death all over the world, with antimi- crobial resistance presenting a great challenge. New antibiotics need to be developed to provide therapeutic treatment options, requiring novel drug targets to be identified and pursued. DNA topoi- somerases control the topology of DNA via DNA cleavage–rejoining coupled to DNA strand passage. The change in DNA topological features must be controlled in vital processes including DNA repli- cation, transcription, and DNA repair. Type IIA topoisomerases are well established targets for antibiotics. In this review, type IA topoisomerases in bacteria are discussed as potential targets for new antibiotics. In certain bacterial pathogens, topoisomerase I is the only type IA topoisomerase present, which makes it a valuable antibiotic target. This review will summarize recent attempts that have been made to identify inhibitors of bacterial topoisomerase I as potential leads for antibiotics and use of these inhibitors as molecular probes in cellular studies. Crystal structures of inhibitor–enzyme complexes and more in-depth knowledge of their mechanisms of actions will help to establish the structure–activity relationship of potential drug leads and develop potent and selective therapeutics that can aid in combating the drug resistant bacterial infections that threaten public health. -

Fusion Transcripts and Transcribed Retrotransposed Loci Discovered Through Comprehensive Transcriptome Analysis Using Paired-End Ditags (Pets)
Downloaded from genome.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs) Yijun Ruan,1,6 Hong Sain Ooi,2 Siew Woh Choo,2 Kuo Ping Chiu,2 Xiao Dong Zhao,1 K.G. Srinivasan,1 Fei Yao,1 Chiou Yu Choo,1 Jun Liu,1 Pramila Ariyaratne,2 Wilson G.W. Bin,2 Vladimir A. Kuznetsov,2 Atif Shahab,3 Wing-Kin Sung,2,4 Guillaume Bourque,2 Nallasivam Palanisamy,5 and Chia-Lin Wei1,6 1Genome Technology and Biology Group, Genome Institute of Singapore, Singapore 138672, Singapore; 2Information and Mathematical Science Group, Genome Institute of Singapore, Singapore 138672, Singapore; 3Bioinformatics Institute, Singapore 138671, Singapore; 4School of Computing, National University of Singapore, Singapore 117543, Singapore; 5Cancer Biology Group, Genome Institute of Singapore, Singapore 138672, Singapore Identification of unconventional functional features such as fusion transcripts is a challenging task in the effort to annotate all functional DNA elements in the human genome. Paired-End diTag (PET) analysis possesses a unique capability to accurately and efficiently characterize the two ends of DNA fragments, which may have either normal or unusual compositions. This unique nature of PET analysis makes it an ideal tool for uncovering unconventional features residing in the human genome. Using the PET approach for comprehensive transcriptome analysis, we were able to identify fusion transcripts derived from genome rearrangements and actively expressed retrotransposed pseudogenes, which would be difficult to capture by other means. Here, we demonstrate this unique capability through the analysis of 865,000 individual transcripts in two types of cancer cells. -
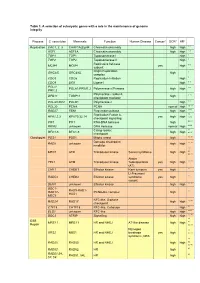
Table 1. a Selection of Eukaryotic Genes with a Role in the Maintenance of Genome Integrity
Table 1. A selection of eukaryotic genes with a role in the maintenance of genome integrity Process S. cerevisiae Mammals Function Human Disease Cancer* GCR* HR* Replication CAC1, 2, 3 CHAF1A,B,p48 Chromatin assembly high high 1, 2 1, 2 ASF1 ASF1A Chromatin assembly high high 3 TOP1 TOP1 Topoisomerase I high 3 TOP2 TOP2 Topoisomerase II high Replicative helicase 4, 5 MCM4 MCM4 yes high subunit Origin Replication 6 ORC3/5 ORC3/6L high complex 7 CDC6 CDC6 Replication Initiation high 7, 8 CDC9 LIG1 Ligase I high POL1/ 7-10 POLA1/PRIM1,2 Polymerase α/Primase high high PRI1,2 Polymerase ε subunit, 6, 11 DPB11 TOBP11 high checkpoint mediator POL3/CDC2 POLD1 Polymerase δ high 7, 8 POL30 PCNA PCNA normal high 12, 13 RAD27 FEN1 Flap endonuclease high high 14-16 Replication Factor A, 14, RFA1,2,3 RPA70,32,14 yes high high 17-19 checkpoint signalling PIF1 PIF1 RNA-DNA helicase high 20, 21 RRM3 unknown DNA Helicase normal high 22-26 Clamp loader, 11, RFC1-5 RFC1-5 high high 20, 27 checkpoint Checkpoint PDS1 PDS1 Mitotic arrest high 11, 28 Damage checkpoint 11, 29 RAD9 unknown high high mediator 11, MEC1 ATR Transducer kinase Seckel syndrome high high 28, 30, 31 Ataxia TEL1 ATM Transducer kinase Telangiectasia yes high high 11, 28 (AT) CHK1 CHEK1 Effector kinase Rare tumours yes high 11 Li-Fraumeni RAD53 CHEK2 Effector kinase syndrome yes high 11 variant DUN1 unknown Effector kinase high high 11, 32 DDC1- RAD9-RAD1- 11 RAD17- PCNA-like complex high HUS1 MEC3 RFC-like, S-phase 11, 33 RAD24 RAD17 high high checkponit 34 CTF18 CHTF18 RFC-like, Cohesion -

Supplementary Information – Postema Et Al., the Genetics of Situs Inversus Totalis Without Primary Ciliary Dyskinesia
1 Supplementary information – Postema et al., The genetics of situs inversus totalis without primary ciliary dyskinesia Table of Contents: Supplementary Methods 2 Supplementary Results 5 Supplementary References 6 Supplementary Tables and Figures Table S1. Subject characteristics 9 Table S2. Inbreeding coefficients per subject 10 Figure S1. Multidimensional scaling to capture overall genomic diversity 11 among the 30 study samples Table S3. Significantly enriched gene-sets under a recessive mutation model 12 Table S4. Broader list of candidate genes, and the sources that led to their 13 inclusion Table S5. Potential recessive and X-linked mutations in the unsolved cases 15 Table S6. Potential mutations in the unsolved cases, dominant model 22 2 1.0 Supplementary Methods 1.1 Participants Fifteen people with radiologically documented SIT, including nine without PCD and six with Kartagener syndrome, and 15 healthy controls matched for age, sex, education and handedness, were recruited from Ghent University Hospital and Middelheim Hospital Antwerp. Details about the recruitment and selection procedure have been described elsewhere (1). Briefly, among the 15 people with radiologically documented SIT, those who had symptoms reminiscent of PCD, or who were formally diagnosed with PCD according to their medical record, were categorized as having Kartagener syndrome. Those who had no reported symptoms or formal diagnosis of PCD were assigned to the non-PCD SIT group. Handedness was assessed using the Edinburgh Handedness Inventory (EHI) (2). Tables 1 and S1 give overviews of the participants and their characteristics. Note that one non-PCD SIT subject reported being forced to switch from left- to right-handedness in childhood, in which case five out of nine of the non-PCD SIT cases are naturally left-handed (Table 1, Table S1). -

Identification of Gastric Cancer–Related Genes Using a Cdna Microarray Containing Novel Expressed Sequence Tags Expressed in Gastric Cancer Cells
Vol. 11, 473–482, January 15, 2005 Clinical Cancer Research 473 Identification of Gastric Cancer–Related Genes Using a cDNA Microarray Containing Novel Expressed Sequence Tags Expressed in Gastric Cancer Cells Jeong-Min Kim,1,5 Ho-Yong Sohn,4 overexpressed in z68% of tissues and the MT2A gene Sun Young Yoon,1 Jung-Hwa Oh,1 Jin Ok Yang,1 was down-expressed in 72% of the tissues. Western blotting and immunohistochemical analyses for CDC20 and SKB1 Joo Heon Kim,2 Kyu Sang Song,3 Seung-Moo Rho,2 1 1 5 showed overexpression and localization changes of the Hyan Sook Yoo, Yong Sung Kim, Jong-Guk Kim, corresponding protein in human gastric cancer tissues. 1 and Nam-Soon Kim Conclusions: Novel genes that are related to human 1Genome Research Center, Korea Research Institute of Bioscience and gastric cancer were identified using cDNA microarray Biotechnology; 2Department of Pathology, Eulji University School of 3 developed in our laboratory. In particular, CDC20 and Medicine; and Department of Pathology, College of Medicine, MT2A represent a potential biomarker of human gastric Chungnam National University, Daejeon, Korea; 4Department of Food and Nutrition, Andong National University, Andong, Korea; and cancer. These newly identified genes should provide a 5Department of Microbiology, College of Natural Sciences, Kyungpook valuable resource for understanding the molecular mecha- National University, Daegu, Korea nism associated with tumorigenesis of gastric carcinogenesis and for the discovery of potential diagnostic markers of gastric cancer. ABSTRACT Purpose: Gastric cancer is one of the most frequently INTRODUCTION diagnosed malignancies in the world, especially in Korea and Japan. -

Two Closely Related Recq Helicases Have Antagonistic Roles in Homologous Recombination and DNA Repair in Arabidopsis Thaliana
Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana Frank Hartung, Stefanie Suer, and Holger Puchta* Botany II, University Karlsruhe, Kaiserstrasse 12, D-76128 Karlsruhe, Germany Edited by Nina Fedoroff, U.S. Department of State, Washington, DC, and approved October 4, 2007 (received for review June 26, 2007) RecQ helicases are involved in the processing of DNA structures RMI1 protein was characterized in mammals and yeast (10, 18, arising during replication, recombination, and repair throughout all 19). Moreover, in the lower eukaryotes S. cerevisiae and Schizo- kingdoms of life. Mutations of different RecQ homologues are re- saccharomyces pombe deletion of their respective RecQ homo- sponsible for severe human diseases, such as Blooms (BLM) or Werner logue leads to partial suppression of the severe phenotypes (WRN) syndrome. The loss of RecQ function is often accompanied by caused by mutation of the TOP3 gene (14, 20–22). hyperrecombination caused by a lack of crossover suppression. In the Several RecQ helicase mutants are synthetically lethal in a Arabidopsis genome seven different RecQ genes are present. Two of combination with mutations in the endonuclease MUS81 (9, 23, them (AtRECQ4A and 4B) arose because of a recent duplication and 24). This inviability of the double mutants is most probably because are still nearly 70% identical on a protein level. Knockout of these both proteins act in parallel pathways resolving aberrant DNA genes leads to antagonistic phenotypes: the RECQ4A mutant shows structures arising during replication. When both genes are mutated, sensitivity to DNA-damaging agents, enhanced homologous recom- these structures accumulate, leading to cell death (9, 23). -

(12) United States Patent (10) Patent No.: US 8,148,129 B2 Frankel Et Al
US008148129B2 (12) United States Patent (10) Patent No.: US 8,148,129 B2 Frankel et al. (45) Date of Patent: Apr. 3, 2012 (54) GENERATION OF POTENT DOMINANT 6,824,978 B1 1 1/2004 Cox, III et al. NEGATIVE TRANSCRIPTIONAL 6,933,113 B2 8, 2005 Case et al. 6,979,539 B2 12/2005 Cox, III et al. INHIBITORS 7,013,219 B2 3/2006 Case et al. 7,070,934 B2 7/2006 Cox, III et al. (75) Inventors: Alan Frankel, Mill Valley, CA (US); 7,163,824 B2 1/2007 Cox, III et al. Robert Nakamura, San Francisco, CA 7,220,719 B2 5/2007 Case et al. (US); Chandreyee Das, Brookline, MA 7,235,354 B2 6/2007 Case et al. 7,262,054 B2 8/2007 Jamieson et al. (US); Ivan D’Orso, San Francisco, CA 7,273,923 B2 9/2007 Jamieson et al. (US); Jocelyn Grunwell, San Mateo, 2003, OO82552 A1* 5, 2003 Wolffe et al. ..................... 435/6 CA (US) (73) Assignee: The Regents of the University of OTHER PUBLICATIONS California, Oakland, CA (US) Cramer et al., Coupling of Transcription with Alternative Splicing: RNA Pol II Promoters Modulate SF2. ASF and 9G8 Effects on an (*) Notice: Subject to any disclaimer, the term of this Exonic Splicing Enhancer, Molecular Cell, 1999, 4:251-258.* patent is extended or adjusted under 35 Cama-Carvalho et al., Nucleocytoplasmic shuttling of heterodimeric U.S.C. 154(b) by 806 days. splicing factor U2AF, JBC. Published on Dec. 15, 2000 as Manu script M008759200.* (21) Appl. No.: 11/765,592 Rosonina et al., Gene Expression: The Close Coupling of Transcrip tion and Splicing, Current Biology, vol. -

Rare Coding Variants in Five DNA Damage Repair Genes Associate
medRxiv preprint doi: https://doi.org/10.1101/2021.04.18.21255506; this version posted April 20, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . 1 Rare coding variants in five DNA damage repair genes 2 associate with timing of natural menopause 3 Lucas D. Ward1*, Margaret M. Parker1, Aimee M. Deaton1, Ho-Chou Tu1, Alexander O. Flynn-Carroll1, 4 Gregory Hinkle1, Paul Nioi1 5 6 1. Alnylam Pharmaceuticals, Cambridge, MA 02142, USA. 7 * To whom correspondence should be addressed. Email: [email protected]. 8 9 Abstract 10 The age of menopause is associated with fertility and disease risk, and its genetic control is of great 11 interest. We used whole-exome sequences from 119,992 women in the UK Biobank to test for 12 associations between rare damaging variants and age at natural menopause. Rare damaging variants in 13 three genes significantly associated with menopause: CHEK2 (p = 6.2 x 10-51) and DCLRE1A (p = 1.2 x 14 10-12) with later menopause and TOP3A (p = 8.8 x 10-8) with earlier menopause. Two additional genes 15 were suggestive: RAD54L (p = 2.3 x 10-6) with later menopause and HROB (p = 2.7 x 10-6) with earlier 16 menopause. In a follow-up analysis of repeated questionnaires in women who were initially pre- 17 menopausal, CHEK2, TOP3A, and RAD54L genotype associated with subsequent menopause. -

SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability
International Journal of Molecular Sciences Review SUMO-Targeted Ubiquitin Ligases and Their Functions in Maintaining Genome Stability Ya-Chu Chang † , Marissa K. Oram † and Anja-Katrin Bielinsky * Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, Minnesota, MN 55455, USA; [email protected] (Y.-C.C.); [email protected] (M.K.O.) * Correspondence: [email protected]; Tel.: +1-612-624-2469 † These authors contributed equally. Abstract: Small ubiquitin-like modifier (SUMO)-targeted E3 ubiquitin ligases (STUbLs) are special- ized enzymes that recognize SUMOylated proteins and attach ubiquitin to them. They therefore connect the cellular SUMOylation and ubiquitination circuits. STUbLs participate in diverse molec- ular processes that span cell cycle regulated events, including DNA repair, replication, mitosis, and transcription. They operate during unperturbed conditions and in response to challenges, such as genotoxic stress. These E3 ubiquitin ligases modify their target substrates by catalyzing ubiquitin chains that form different linkages, resulting in proteolytic or non-proteolytic outcomes. Often, STUbLs function in compartmentalized environments, such as the nuclear envelope or kine- tochore, and actively aid in nuclear relocalization of damaged DNA and stalled replication forks to promote DNA repair or fork restart. Furthermore, STUbLs reside in the same vicinity as SUMO proteases and deubiquitinases (DUBs), providing spatiotemporal control of their targets. In this review, we focus on the molecular mechanisms by which STUbLs help to maintain genome stability Citation: Chang, Y.-C.; Oram, M.K.; across different species. Bielinsky, A.-K. SUMO-Targeted Ubiquitin Ligases and Their Keywords: genome stability; STUbL; SUMO; ubiquitin; Slx5/Slx8; RNF4; RNF111 Functions in Maintaining Genome Stability. -

Characterization of Recombinant Human DNA Topoisomerase Iiiƒї
J Biochem. 127. 1109-1113(2000) Characterization of Recombinant Human DNA Topoisomerase IIIƒ¿ Activity Expressed in Yeast1 Nika Hotoda2 and Ryo Hanai3 Department of Chemistry , College of Science,Rikkyo (St. Paul's) Unioersity,3-34-1 Nishi-Ikebukure, Toshirna-ku, Tokyo171-8501 Received January 28.2000; accepted April 3, 2000 Recombinant human DNA topoisomerase IIIƒ¿ was expressed in mutant yeast cells devoid of both topoisomerases I and III, and the gene product was partially purified. The activity of the protein in supercoil removal was found to be limited and also bipha sic: in the first phase it processively changed the linking-number of hypernegatively supercoiled DNA, but only to the superhelicity of a regular negative supercoil; in the second phase the enzyme relaxed the DNA further, but only slightly and slowly. The optimal solution conditions for the enzyme activity were found to be physiological. The assay results with a truncation mutant showed that the C-terminal 334 amino acids are unnecessary for the activity, suggesting that this region, and perhaps the entire protein, is involved in a function other than supercoil removal. Key words: DNA supereoiling, DNA topoisomerase. Mammalian DNA topoisomerase III (TOP3 gene product), merase I (for a review, 14), but absent in yeast topoiso a type-IA topoisomerase, was first identified in man (I), merase III. Although bacterial topoisomerase I can relax and has been shown to be essential for embryonic develop negatively supercoiled DNA nearly completely, eukaryotic ment in the mouse (2). More recently, a second TOP3 gene type-IA topoisomerases studied so far (1, 4, 12), including was identified (3-5), and the first gene has been renamed the yeast enzyme, appear to remove negative supercoils TOP3ƒ¿ and the second one designated as TOP3ƒÀ (2).