Acute Promyelocytic Leukemia: a Constellation of Molecular Events Around a Single PML-RARA Fusion Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Minimal Residual Disease Detection in Childhood Precursor–B-Cell Acute Lymphoblastic Leukemia: Relation to Other Risk Factors
Leukemia (2003) 17, 1566–1572 & 2003 Nature Publishing Group All rights reserved 0887-6924/03 $25.00 www.nature.com/leu Minimal residual disease detection in childhood precursor–B-cell acute lymphoblastic leukemia: relation to other risk factors. A Children’s Oncology Group study MJ Borowitz1, DJ Pullen2, JJ Shuster3,4, D Viswanatha5, K Montgomery6, CL Willman5 and B Camitta7 1Johns Hopkins Medical Institutions, USA; 2University of Mississippi, USA; 3University of Florida, Gainesville FL, USA; 4Children’s Oncology Group, Arcadia, CA, USA; 5University of New Mexico, USA; 6Genzyme Genetics, Santa Fe, NM; and 7Milwaukee Childrens Hospital, USA Minimal residual disease (MRD) can be detected in the marrows prognostic significance at several time points. MRD levels at the of children undergoing chemotherapy either by flow cytometry end of induction are highly prognostic. Since intensification of or polymerase chain reaction. In this study, we used four-color therapy after successful induction can improve the outcome of flow cytometry to detect MRD in 1016 children undergoing 33 therapy on Children’s Oncology Group therapeutic protocols many patients with adverse prognostic features, it is possible for precursor–B-cell ALL. Compliance was excellent, with that a similar approach might benefit patients with MRD. For follow-up samples received at the end of induction on nearly these reasons, we have chosen to focus our MRD studies on 95% of cases; sensitivity of detection at this time point was at end-of-induction therapy. least 1/10,000 in more than 90% of cases. Overall, 28.6% of Although MRD and other measures of early response to patients had detectable MRD at the end of induction. -

FIP1L1 Gene FIP1 Like 1 (S
FIP1L1 gene FIP1 like 1 (S. cerevisiae) Normal Function The FIP1L1 gene provides instructions for making part of a protein complex named cleavage and polyadenylation specificity factor (CPSF). This complex of proteins plays an important role in processing molecules called messenger RNAs (mRNAs), which serve as the genetic blueprints for making proteins. The CPSF protein complex helps add a string of the RNA building block adenine to the mRNA, creating a polyadenine tail or poly(A) tail. The poly(A) tail is important for stability of the mRNA and for protein production from the blueprint. Health Conditions Related to Genetic Changes PDGFRA-associated chronic eosinophilic leukemia A deletion of genetic material from chromosome 4 brings together part of the FIP1L1 gene and part of another gene called PDGFRA, creating the FIP1L1-PDGFRA fusion gene. This mutation is a somatic mutation, which means it is acquired during a person's lifetime and is present only in certain cells. This fusion gene causes PDGFRA- associated chronic eosinophilic leukemia, which is a type of blood cell cancer characterized by an increased number of eosinophils, a type of white blood cell involved in allergic reactions. The FIP1L1-PDGFRA protein produced from the fusion gene has the function of the normal PDGFRA protein, which stimulates signaling pathways inside the cell that control many important cellular processes, such as cell growth and division (proliferation) and cell survival. Unlike the normal PDGFRA protein, however, the FIP1L1-PDGFRA protein is constantly turned on (constitutively activated), which means the cells are always receiving signals to proliferate. When the FIP1L1-PDGFRA fusion gene occurs in blood cell precursors, the growth of eosinophils (and occasionally other blood cells) is poorly controlled, leading to PDGFRA-associated chronic eosinophilic leukemia. -

R-Loop-Mediated Genome Instability in Mrna Cleavage and Polyadenylation Mutants
Downloaded from genesdev.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants Peter C. Stirling,1,3 Yujia A. Chan,1,3 Sean W. Minaker,1,3 Maria J. Aristizabal,2 Irene Barrett,1 Payal Sipahimalani,1 Michael S. Kobor,2 and Philip Hieter1,4 1Michael Smith Laboratories, University of British Columbia, Vancouver, British Columbia V6T1Z4, Canada; 2Centre for Molecular Medicine and Therapeutics, Vancouver, British Columbia V5Z4H4, Canada Genome instability via RNA:DNA hybrid-mediated R loops has been observed in mutants involved in various aspects of transcription and RNA processing. The prevalence of this mechanism among essential chromosome instability (CIN) genes remains unclear. In a secondary screen for increased Rad52 foci in CIN mutants, representing ~25% of essential genes, we identified seven essential subunits of the mRNA cleavage and polyadenylation (mCP) machinery. Genome-wide analysis of fragile sites by chromatin immunoprecipitation (ChIP) and microarray (ChIP–chip) of phosphorylated H2A in these mutants supported a transcription-dependent mechanism of DNA damage characteristic of R loops. In parallel, we directly detected increased RNA:DNA hybrid formation in mCP mutants and demonstrated that CIN is suppressed by expression of the R-loop-degrading enzyme RNaseH. To investigate the conservation of CIN in mCP mutants, we focused on FIP1L1, the human ortholog of yeast FIP1, a conserved mCP component that is part of an oncogenic fusion in eosinophilic leukemia. We found that truncation fusions of yeast FIP1 analogous to those in cancer cause loss of function and that siRNA knockdown of FIP1L1 in human cells increases DNA damage and chromosome breakage. -

Hypereosinophilia in Granular Acute Bcell Lymphoblastic Leukemia With
bs_bs_banner Microscopic polyangiitis 543 and had been diagnosed with normocytic anemia.At that time, she References received treatment with an iron preparation because of a low 1 Ozen S, Ruperto N, Dillon MJ et al. EULAR/PReS endorsed con- serum iron level despite her normal ferritin level. We estimate that sensus criteria for the classification of childhood vasculitides. Ann. the patient already had mild alveolar hemorrhage at that time and Rheum. Dis. 2006; 65: 936–41. the bleeding later healed spontaneously. Among the cases of this 2 Vanoni F, Bettinelli A, Keller F, Bianchetti MG, Simonetti GD. disease reported from Japan, the disease tended to develop more Vasculitides associated with IgG antineutrophil cytoplasmic autoan- tibodies in childhood. Pediatr. Nephrol. 2010; 25: 205–12. frequently in the spring. In our patient, the disease developed twice 3 Kobayashi S, Inokuma S. Intrapulmonary hemorrhage in collagen- (both times in June), suggesting the possibility that infection vascular disease includes a spectrum of underlying conditions. triggered the disease onset. Although alveolar hemorrhage sub- Intern. Med. 2009; 48: 891–7. sided spontaneously in this case, lung fibrosis may develop in the 4 Hattori M, Kurayama H, Koitabashi Y. Antineutrophil cytoplasmic future if alveolar bleeding develops repeatedly and its diagnosis is autoantibody-associated glomerulonephritis in children. J. Am. Soc. Nephrol. 2001; 12: 1493–500. delayed. In the present case, treatment was started immediately 5 Peco-Antic A, Bonaci-Nikolic B, Basta-Jovanovic G et al. Child- after diagnosis and rapid aggravation of her renal function was hood microscopic polyangiitis associated with MPO-ANCA. prevented. When dealing with cases in whom unexplained nor- Pediatr. -

WO 2017/180694 Al 19 October 2017 (19.10.2017) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2017/180694 Al 19 October 2017 (19.10.2017) P O P C T (51) International Patent Classification: DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, C12N 15/10 (2006.01) C12N 15/63 (2006.01) HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KH, KN, C12N 15/62 (2006.01) C12N 9/22 (2006.01) KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, (21) International Application Number: NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, PCT/US20 17/027 126 RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, (22) International Filing Date: TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, 12 April 2017 (12.04.2017) ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, (30) Priority Data: TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, 62/322,026 13 April 2016 (13.04.2016) US TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, (71) Applicant: EDITAS MEDICINE, INC. -

PRODUCT SPECIFICATION Prest Antigen FIP1L1 Product Datasheet
PrEST Antigen FIP1L1 Product Datasheet PrEST Antigen PRODUCT SPECIFICATION Product Name PrEST Antigen FIP1L1 Product Number APrEST79770 Gene Description factor interacting with PAPOLA and CPSF1 Alternative Gene DKFZp586K0717 Names Corresponding Anti-FIP1L1 (HPA037475) Antibodies Description Recombinant protein fragment of Human FIP1L1 Amino Acid Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: KANSSVGKWQDRYGRAESPDLRRLPGAIDVIGQTITISRVEGRRRANENS NIQVLSERSATEVDNNFSKPPPFFPPGAPPT Fusion Tag N-terminal His6ABP (ABP = Albumin Binding Protein derived from Streptococcal Protein G) Expression Host E. coli Purification IMAC purification Predicted MW 27 kDa including tags Usage Suitable as control in WB and preadsorption assays using indicated corresponding antibodies. Purity >80% by SDS-PAGE and Coomassie blue staining Buffer PBS and 1M Urea, pH 7.4. Unit Size 100 µl Concentration Lot dependent Storage Upon delivery store at -20°C. Avoid repeated freeze/thaw cycles. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -
Letters to the Editor
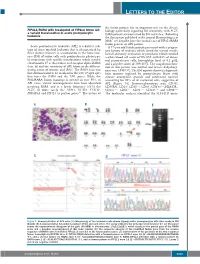
LETTERS TO THE EDITOR the fusion partner has an important role on the disease FIP1L1/RARA with breakpoint at FIP1L1 intron 13: biology particularly regarding RA sensitivity, with PLZF- a variant translocation in acute promyelocytic 4 leukemia RARA patients characterized by RA resistance. Following the description published in the journal Haematologica in 2008, 5 we describe here the second case of FIP1L1/RARA fusion gene in an APL patient. Acute promyelocytic leukemia (APL) is a distinct sub - A 77-year old female patient presented with a progres - type of acute myeloid leukemia that is characterized by sive history of asthenia which lasted for several weeks. three distinct features: i) accumulation in the bone mar - Initial laboratory evaluation of peripheral blood revealed row (BM) of tumor cells with promyelocytic phenotype; a white blood cell count of 59 ¥10 9/L with 84% of abnor - ii) association with specific translocations which involve mal promyelocyte cells, hemoglobin level of 9.2 g/dL, chromosome 17 at the retinoic acid receptor alpha ( RARA ) and a platelet count of 109 ¥10 9/L. The coagulation func - locus ; iii) and the sensitivity of APL blasts to the differen - tion in this patient was normal and lactate dehydroge - 1 tiating action of retinoic acid (RA). The RARA locus was nase was 1,938 U/L. The BM aspirate showed a hypercel - first demonstrated to be involved in the t(15;17)(q22;q21) lular marrow replaced by promyelocyte blasts with that fuses the RARA and the PML genes. While the intense azurophilic granule and prominent nucleoli PML/RARA fusion transcript is present in over 95% of accounting for 93% of all nucleated cells, suggestive of APL cases, variant rearrangements have been identified APL (Figure 1A). -

Effect of the Philadelphia Chromosome on Minimal Residual Disease In
Leukemia (1997) 11, 1497–1500 1997 Stockton Press All rights reserved 0887-6924/97$12.00 Effect of the Philadelphia chromosome on minimal residual disease in acute lymphoblastic leukemia MJ Brisco1, PJ Sykes1, G Dolman1, S-H Neoh1, E Hughes1, L-M Peng2, G Tauro3, H Ekert3, I Toogood4, K Bradstock5 and AA Morley1 1Department of Haematology, Flinders Medical Centre, Bedford Park, South Australia; 2Department of Laboratory Medicine, School of Medicine, West China University of Medical Sciences, Chengdu, People’s Republic of China; 3Department of Haematology, Royal Childrens Hospital, Parkville, Victoria; 4Department of Haematology/Oncology, Womens and Childrens Hospital, North Adelaide, South Australia; and 5Department of Haematology, Westmead Hospital, Westmead, NSW, Australia The Philadelphia translocation is associated with a poor prog- number of leukemic cells remaining at the end of induction nosis in adults and children with acute lymphoblastic leukemia, provides a good approximation of the degree of drug resist- even though the majority of patients achieve remission. To test ance in vivo. the hypothesis that the translocation leads to drug resistance 6–10 in vivo, we studied 61 children and 20 adults with acute lym- Since the initial reports, a number of groups have phoblastic leukemia and used the level of minimal residual dis- developed sensitive methods to quantify minimal residual dis- ease at the end of induction as the measure of drug resistance ease (MRD) by use of the polymerase chain reaction (PCR). in vivo. In children -

REVIEW Clinical Relevance of Minimal Residual Disease Monitoring in Non
Leukemia (1999) 13, 1691–1695 1999 Stockton Press All rights reserved 0887-6924/99 $15.00 http://www.stockton-press.co.uk/leu REVIEW Clinical relevance of minimal residual disease monitoring in non-Hodgkin’s lymphomas: a critical reappraisal of molecular strategies P Corradini1, M Ladetto2, A Pileri2 and C Tarella2 1Bone Marrow Transplantation Unit, Istituto Scientifico HS Raffaele, Milan; and 2Divisione Universitaria di Ematologia–Azienda Ospedaliera S Giovanni Battista, Turin, Italy Although current treatments can induce clinical complete neoplasms. In the NHL setting, several studies have been pub- remissions in the vast majority of patients with indolent lym- lished on the prognostic significance of minimal residual dis- phoma, most of them actually relapse, because of the persist- ence of residual tumor cells which are undetectable using con- ease (MRD) detection. Most of these studies focus on indolent ventional diagnostic procedures. Polymerase chain reaction lymphomas: small lymphocytic lymphoma/chronic lympho- (PCR)-based methods are increasingly used for minimal cytic leukemia (SLL/CLL),17,18 follicular (FCL)19–21 and mantle residual disease detection (MRD), and provide useful prognos- cell lymphomas (MCL).20,22 This is mostly because these tic information. In this review, current approaches for MRD tumors, unlike more aggressive NHL histotypes, are dissemi- detection in indolent lymphomas are summarized. In addition, nated disorders with frequent microscopic or submicroscopic the prognostic aspects of molecular monitoring after transplan- tation procedures are discussed. The experience accumulated bone marrow (BM) and peripheral blood (PB) involvement, over the past decade shows that PCR analysis has a prognostic and thus they represent ideal targets for MRD evaluation with impact in several therapeutic programs including conventional PCR-based assays.23,24 and high-dose regimens. -

ORIGINAL ARTICLE the Severity of FIP1L1–PDGFRA-Positive Chronic
Leukemia (2007) 21, 2428–2432 & 2007 Nature Publishing Group All rights reserved 0887-6924/07 $30.00 www.nature.com/leu ORIGINAL ARTICLE The severity of FIP1L1–PDGFRA-positive chronic eosinophilic leukaemia is associated with polymorphic variation at the IL5RA locus S Burgstaller1, S Kreil1, K Waghorn1, G Metzgeroth2, C Preudhomme3, K Zoi4, H White1, D Cilloni5, C Zoi4, F Brito-Babapulle6, C Walz2, A Reiter2 and NCP Cross1 1Wessex Regional Genetics Laboratory, University of Southampton, Salisbury, UK; 2III Medizinische Universita¨tsklinik, Medizinische Fakulta¨t der Universita¨t Mannheim, Heidelberg, Germany; 3Laboratoire d’Hematologie A, CHU Lille, Lille, France; 4Foundation of Biomedical Research, Academy of Athens, Athens, Greece; 5Department of Clinical and Biological Sciences, University of Turin, Turin, Italy and 6Department of Haematology, Ealing Hospital, London, UK We have investigated the hypothesis that constitutional genetic Understanding of the myeloproliferative subtype of HES was variation in IL-5 signalling may be associated with the greatly advanced by the finding of the FIP1L1–PDGFRA fusion, development or severity of FIP1L1–PDGFRA-positive chronic formed by a cytogenetically cryptic 800 kb interstitial deletion at eosinophilic leukaemia (CEL) in humans. We genotyped six 2 single-nucleotide polymorphisms (SNP) within or close to the chromosome band 4q12. Although initially described in more IL5RA or IL5 genes in 82 patients with FIP1L1–PDGFRA- than 50% of cases with idiopathic HES, subsequent studies positive CEL plus, as controls, healthy individuals (n ¼ 100), estimated the prevalence to be lower at 3À17%.3–5 Detection of patients with FIP1L1–PDGFRA-negative eosinophilia (n ¼ 100) FIP1L1–PDGFRA by reverse transcription-PCR or fluorescent in or patients with chronic myeloid leukaemia (CML) (n ¼ 100). -

How Close Are We to Incorporating Measurable Residual Disease Into
REVIEW ARTICLE How close are we to incorporating Ferrata Storti Foundation measurable residual disease into clinical practice for acute myeloid leukemia? Nicholas J. Short and Farhad Ravandi Department of Leukemia, The University of Texas MD Anderson Cancer Center, Houston, TX, USA Haematologica 2019 ABSTRACT Volume 104(8):1532-1541 ssessment of measurable residual disease, also called “minimal resid - ual disease,” in patients with acute myeloid leukemia in morpholog - Aical remission provides powerful prognostic information and comple - ments pretreatment factors such as cytogenetics and genomic alterations. Based on data that low levels of persistent or recurrent residual leukemia are consistently associated with an increased risk of relapse and worse long- term outcomes, its routine assessment has been recommended by some experts and consensus guidelines. In addition to providing important prog - nostic information, the detection of measurable residual disease may also theoretically help to determine the optimal post-remission strategy for an individual patient. However, the full therapeutic implications of measurable residual disease are uncertain and thus controversy exists as to whether it should be routinely incorporated into clinical practice. While some evidence supports the use of allogeneic stem cell transplantation or hypomethylating agents for some subgroups of patients in morphological remission but with detectable residual leukemia, the appropriate use of this information in mak - ing clinical decisions remains largely speculative at present. To resolve this pressing clinical issue, several ongoing studies are evaluating measurable Correspondence: residual disease-directed treatments in acute myeloid leukemia and may FARHAD RAVANDI lead to new, effective strategies for patients in these circumstances. This [email protected] review examines the common technologies used in clinical practice and in the research setting to detect residual leukemia, the major clinical studies Received: February 8, 2019. -

FIP1L1‑PDGFRA Fusion‑Negative Hypereosinophilic Syndrome with Uncommon Cardiac Involvement Responding to Imatinib Treatment: a Case Report
MOLECULAR AND CLINICAL ONCOLOGY 9: 35-39, 2018 FIP1L1‑PDGFRA fusion‑negative hypereosinophilic syndrome with uncommon cardiac involvement responding to imatinib treatment: A case report AMANDA SANTOS DAL BERTO1, RICARDO HOHMANN CAMIÑA1, EDUARDO SILVA MACHADO1-3 and ANTUANI RAFAEL BAPTISTELLA1,2,4,5 1Santa Terezinha University Hospital; 2University of West Santa Catarina; 3Department of Clinical Oncology, Santa Terezinha University Hospital; 4Oncology Research Group of Santa Terezinha University Hospital /University of West Santa Catarina; 5Post Graduation Program in Bioscience and Health/University of West Santa Catarina, Joaçaba, Santa Catarina 89600-000, Brazil Received April 17, 2018; Accepted May 21, 2018 DOI: 10.3892/mco.2018.1637 Abstract. Hypereosinophilic syndrome is a rare, chronic therapeutic possibilities were evaluated due to the significant hematological disease characterized by a persistently elevated progression of the disease, and it was decided to attempt the eosinophil count exceeding 1.5x109/l, following the exclusion use of imatinib, despite its use being preferably recommended of other potential etiologies. The systemic involvement of the for FIPIL1-PDGFRA-positive patients. The patient exhibited an disease causes tissue damage through eosinophil infiltration, evident and immediate response to imatinib, with normalization and may affect various organs; cardiac complications are of the eosinophil count within 24 h of the first dose, which was observed in 50-60% of cases, which are predominately attrib- maintained for at least the next 19 months. This clinical presen- uted to endomyocardial fibrosis. The treatment is based initially tation is uncommon, as patients negative for FIPIL1-PDGFRA on determining the presence of the FIP1L1-PDGFRA fusion. fusion do not frequently respond to imatinib treatment, and Patients with positive results for this mutation tend to achieve symptomatic heart failure usually appears in the third stage of a complete response with imatinib treatment, which is thus disease progression.