Microsatellite Markers Linked to Six Russian Wheat Aphid Resistance Genes in Wheat
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Il Calendario Dei Divieti Di Circolazione Della Grecia E Della Spagna Non È Ancora Disponibile
Driving restrictions, 2008 Austria 1. GENERAL DRIVING RESTRICTIONS Vehicles concerned Trucks with trailers, if the maximum authorised total weight of the motor vehicle or the trailer exceeds 3.5t; trucks, articulated vehicles and self-propelled industrial machines with an authorised total weight of more than 7.5t. Area Nationwide, with the exception of journeys made exclusively as part of a combined transport operation within a radius of 65km of the following transloading stations: Brennersee; Graz-Ostbahnhof; Salzburg-Hauptbahnhof; Wels-Verschiebebahnhof; Villach-Fürnitz; Wien-Südbahnhof; Wien-Nordwestbahnhof; Wörg; Hall in Tirol CCT; Bludenz CCT; Wolfurt CCT. Prohibition Saturdays from 15h00 to 24h00; Sundays and public holidays from 00h00 to 22h00 Public holidays 2008 1 January New Year’s Day 6 January Epiphany 24 March Easter Monday 1 May Labour Day; Ascension 12 May Whit Monday 22 May Corpus Christi 15 August Assumption 26 October National holiday 1 November All Saints’ Day 8 December Immaculate Conception 25 December Christmas Day 26 December Boxing Day Exceptions concerning trucks with trailers exceeding 3.5t · vehicles transporting milk; concerning vehicles with an authorised total weight of more than 7.5t · vehicles carrying meat or livestock for slaughter (but not the transport of heavy livestock on motorways), perishable foodstuffs (but not deep frozen goods), the supply of refreshments to tourist areas, urgent repairs to refrigeration plant, towing services (in all cases, according to § 46 StVO, it is obligatory to leave the motorway at the nearest exit), breakdown assistance vehicles, emergency vehicles, vehicles of a scheduled transport company (regular lines), and local trips on the two Saturdays preceding 24 December. -
An Empirical Analysis of the Relation Between Infrastructure and Road Accidents
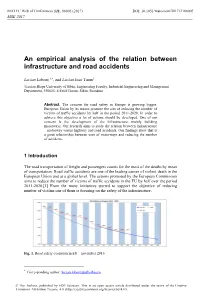
MATEC Web of Conferences 121, 06005 (2017) DOI: 10.1051/ matecconf/201712106005 MSE 2017 An empirical analysis of the relation between infrastructure and road accidents Lucian Lobonț 1,*, and Lucian Ioan Tarnu1 1Lucian Blaga University of Sibiu, Engineering Faculty, Industrial Engineering and Management Department, 550025, 4 Emil Cioran, Sibiu, România Abstract. The concern for road safety in Europe is growing bigger. European Union by its means promote the aim of reducing the number of victims of traffic accidents by half in the period 2011-2020. In order to achieve this objective a lot of actions should be developed. One of our concern is the development of the infrastructure, mainly building motorways. Our research aims to study the relation between infrastructure – motorway versus highway and road accidents. Our findings show that is a great relationship between uses of motorways and reducing the number of accidents. 1 Introduction The road transportation of freight and passengers counts for the most of the deaths by mean of transportation. Road traffic accidents are one of the leading causes of violent death in the European Union and at a global level. The actions promoted by the European Commission aims to reduce the number of victims of traffic accidents in the EU by half over the period 2011-2020.[1] From the many initiatives started to support the objective of reducing number of victims one of them is focusing on the safety of the infrastructure. Fig. 1. Road safety evolution in EU – november 2016 * Corresponding author: [email protected] © The Authors, published by EDP Sciences. This is an open access article distributed under the terms of the Creative Commons Attribution License 4.0 (http://creativecommons.org/licenses/by/4.0/). -

REPORT Board of Directors 2019
REPORT Board of Directors 2019 1 Board of Directors Report for the year 2019 – Impact Group Content 03 2019 Highlights 04 Impact Group, overview 05 Projects portfolio 10 Activity of Impact Group in 2019 14 Residential real estate market 16 Strategy highlights 17 Financial results 19 Board of Directors and Executive Management 21 Main risks and uncertainties 22 Corporate Governance 24 Impact Developer & Contractor on the capital market 26 Reconciliation of IFRS Net Assets vs. EPRA 27 Annexes 27 Implementation of the Corporate Governance Code 34 Other information 35 Impact Group www.impactsa.ro 2 Board of Directors Report for the year 2019 – Impact Group 2019 HIGHLIGHTS Operational Financial IFRS NAV • 352 dwellings sold (30,185 sqm) • 263 pre-sale agreements and reservations on 31 Dec 584 mLEI 2019 estimated as sales in 2020 + 137.2 mLEI (2018: 446.8 mLEI) • Delivery of 192 apartments (14,500 sqm) for Greenfield Baneasa Residence (“Greenfield”) EPRA NAV • Further development of 500 apartments built in the Luxuria Domenii Residence project (“Luxuria”), to be delivered during 2020 839 mLEI • Starting works for the third phase of the Luxuria + 119 mLEI (2018: 720 mLEI) project, 130 apartments that will be delivered in the first half of 2021 Sales • Obtaining the new Zonal Urban Plan (PUZ) new developments in Greenfield Baneasa (Greenfield IV and Greenfield V), which includes Greenfield Plaza (a 157 mLEI commercial and leisure centre) + 56 mLEI (2018: 101 mLEI) • Obtaining the Zonal Urban Plan (PUZ) for the residential project Boreal Plus to -

Colliers International Romania Mid-Year Market Update
H1 20 18 Colliers International Romania Mid-Year Market Update Accelerating success. 1 2 Colliers International Romania H1 Research & Forecast Report | 2018 Content Romania Macro Industrial Retail Update Market Market p. 04 p. 06 p. 08 Office Investment Land Market Market Market p. 10 p. 12 p. 16 Romania macro update In the post-crisis period, Romania has been the most we saw last year cannot be maintained and the anticipated The major challenges for the Romanian economy going successful economic convergence story in this part of slowdown is upon us. Some transitory factors weighed forward remain structural in nature (so more difficult Europe. In fact, if the service-led growth continues at a on GDP, leading to quasi-flat GDP readings in quarter-on- to tackle), like building highways, cutting back red tape pace similar to the post-crisis period, Romania is likely to quarter terms in 4Q17 and 1Q18, which is not something or corruption, increasing population activity rates and surpass Hungary by end-2022 and catch up to Slovakia we considered. The important note here is that due to improving education. Take the labour market for instance: by end-2028 in terms of GDP per capita, adjusted to the way economic growth is calculated and the statistical employers are finding it ever harder to fill in positions (both purchasing power standards (this indicator is widely used relevance of these quarters, it is looking nigh on impossible for white- and blue-collar positions), with unemployment as a proxy for living standards). to achieve an expansion rate above 5% in 2018 (barring near record lows of 4.4%. -

Polymorphic Analysis of Mitochondrial 12S Rrna Gene of Common Sun Skink Eutropis Multifasciata (Reptilia: Squamata: Scincidae) in Central Vietnam
Available online a t www.scholarsresearchlibrary.com Scholars Research Library Annals of Biological Research, 2015, 6 (11):1-10 (http://scholarsresearchlibrary.com/archive.html) ISSN 0976-1233 CODEN (USA): ABRNBW Polymorphic analysis of mitochondrial 12S rRNA gene of common sun skink Eutropis multifasciata (Reptilia: Squamata: Scincidae) in Central Vietnam Ngo Dac Chung 1, Tran Quoc Dung 1* and Ma Phuoc Huyen Thanh An 2 1Faculty of Biology, College of Education, Hue University, Vietnam 2Faculty of Biology, College of Science, Hue University, Vietnam _____________________________________________________________________________________________ ABSTRACT Analysis of 12S rRNA sequences of twenty specimens from Common Sun Skink Eutropis multifasciata in Central Vietnam showed genetic differences among specimens range from 0% (between the specimens H1, QB1, QB2 and DL6; or H2 and QT1; or H9, HT1, DN1 and NA2; or DN7 and DN8) to 1,79% (between the specimens DN2 and DL5). The mitochondrial tree generated from these sequences confirmed the monophyly of all specimens of E. multifasciata and the monophyly of the genus Eutropis. These mitochondrial 12S rRNA sequences of specimens from E. multifasciata (HI, H2, H3, H9, H10, DL5, DL6, QN1, QN2, QN8, NA2, NA3, QB1, QB2, QT1, HT1, DN1, DN7, DN8, and DN2) were deposited in GenBank with accession number KT350390-KT350409, respectively. Keywords: Eutropis multifasciata , Mabuya , mitochondrial 12S rRNA gene, Reptilia, Squamata _____________________________________________________________________________________________ INTRODUCTION The genus Eutropis or the tropical Asian Mabuya belonging to the family Scincidae, ordo Squamata of Reptile, currently consist of around 30 described species distributed predominantly in the Indomalayan (The Indomalayan region is further divided into Indian, Indochinese and Sundaic subregion) [8] and several yet undescribed species occurring from the Middle East to Palau, Oceania [22]. -

Dn10, a New Gene Conferring Resistance to Russian Wheat Aphid Biotype 2 in Iranian Wheat Landrace PI 682675
Published March 15, 2018 RESEARCH Dn10, a New Gene Conferring Resistance to Russian Wheat Aphid Biotype 2 in Iranian Wheat Landrace PI 682675 Genqiao Li, Xiangyang Xu,* Brett F. Carver, Peiguo Guo, and Gary Puterka G. Li, X. Xu, and G. Puterka, Wheat, Peanut and Other Field Crops ABSTRACT Research Unit, USDA-ARS, Stillwater, OK 74075; B.F. Carver, Plant R ussian wheat aphid [RWA, Diuraphis noxia and Soil Sciences Dep., Oklahoma State Univ., Stillwater, OK 74078; (Mordvilko)] is a serious pest of wheat (Triticum P. Guo, International Crop Research Center for Stress Resistance, aestivum L.) and barley (Hordeum vulgare L.) College of Life Sciences, Guangzhou Univ., Guangzhou 510006, China. that causes heavy yield losses in many coun- Received 31 Oct. 2017. Accepted 18 Dec. 2017. *Corresponding author tries, and RWA biotype 2 (RWA2) is virulent to ([email protected]). Assigned to Associate Editor Eric Olson. most RWA resistance genes. The objective of Abbreviations: PCR, polymerase chain reaction; RWA, Russian this study was to characterize a gene for resis- wheat aphid; SSR, simple sequence repeat. tance to RWA2 in Iranian landrace PI 682675, a single-plant selection from PI 624151. F2:3 and F3:4 families derived from cross PI 682675 ´ ussian wheat aphid [RWA, Diuraphis noxia (Mordvilko)], Zhengyou 6 were used to map the resistance Ra serious pest of wheat (Triticum aestivum L.) and barley gene. PI 682675 carries a dominant resistance (Hordeum vulgare L.), is indigenous to southern Russia, Iran, gene, Dn10, flanked by simple sequence repeat Afghanistan, and countries around the Mediterranean Sea (Hewitt markers Xgwm437 and Xwmc488 on chromo- et al., 1984). -

NHS England Yorkshire & Humber Orthodontic Lots and Locations
NHS England Yorkshire & Humber Orthodontic Lots and Locations Total Number Postcodes Total No. of Locations within Postcodes Lot Name Servicing (including UOA's of UOA's (including but not exclusively) but not exclusively) Lots in Lot North Yorkshire & Humber Craven BD20, BD23, BD24 Crosshills, Settle, Skipton, Craven 6500 1 6500 Grassington Harrogate HG1, HG2, HG3, Harrogate, Knaresborough, Ripon, Harrogate 10318 1 10318 HG4, HG5, YO51, Boroughbridge, Marston Moor YO26 Ward Hambleton and DL6, DL7, DL8, Leeming, Leyburn, Thirsk, Hambleton and 8606 1 8606 Richmondshire DL9, DL10, DL11, Northallerton, Richmond, Richmondshire YO7, YO61 Easingwold Scarborough YO11, YO12, Scarborough, Scalby, Seamer Scarborough and and Ryedale YO13, YO14, Ward, East Ayton, Filey, 9682 1 9682 Ryedale YO17, YO18, Hunmanby, Malton, Pickering, YO62, YO21,YO60 Helmsley, Whitby Selby YO8,LS24, LS25, Selby, Sherburn in Elmet, Selby 6500 1 6500 Tadcaster York YO1, YO10, YO19, Acomb, Bishopthorpe, York 10376 1 10376 YO23, YO24, Dunnington, Haxby, Rawcliffe, YO26, YO30, YO32 East Riding - YO15, YO16,YO25, Bridlington, Flamborough, North East HU18, HU10, Holderness Ward, East Wolds and HU11, HU12, Coastal Ward, Hornsea, Mid East Riding 19699 2 9850 HU13, HU14, Holderness Ward, North HU16, HU17, Holderness Ward, Withernsea, HU18, HU19 Hessle, Beverley, Cottingham East Riding - YO25, YO41, Pocklington, Howdenshire Ward, West YO42, YO43, Goole, Hessle, Beverley, 9850 HU10, HU13, Cottingham, Driffield HU14, HU15, HU16, HU17, DN14 Hull East HU1, HU2, HU7, Branshome, Sutton -

Understanding Hadoop Clusters and the Network
Understanding Hadoop Clusters and the Network Part 1. Introduction and Overview Brad Hedlund http://bradhedlund.com http://www.linkedin.com/in/bradhedlund @bradhedlund BRAD HEDLUND .com Hadoop Server Roles Clients Distributed Data Analytics Distributed Data Storage Map Reduce HDFS Secondary Job Tracker Name Node masters Name Node Data Node & Data Node & Data Node & Task Tracker Task Tracker Task Tracker slaves Data Node & Data Node & Data Node & Task Tracker Task Tracker Task Tracker BRAD HEDLUND .com Hadoop Cluster World switch switch switch switch switch switch switch Name Node Job Tracker Secondary NN Client DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT DN + TT Rack 1 Rack 2 Rack 3 Rack 4 Rack N BRAD HEDLUND .com Typical Workflow • Load data into the cluster (HDFS writes) • Analyze the data (Map Reduce) • Store results in the cluster (HDFS writes) • Read the results from the cluster (HDFS reads) Sample Scenario: How many times did our customers type the word “Fraud” into emails sent to customer service? Huge file containing all emails sent File.txt to customer service BRAD HEDLUND .com Writing files to HDFS OK. Write to I want to write Data Nodes Blocks A,B,C of 1,5,6 File.txt File.txt Client Blk A Blk B Blk C Name Node Data Node 1 Data Node 5 Data Node 6 Data Node N Blk A Blk B Blk C • Client consults Name Node • Client writes block directly to one Data Node • Data Nodes replicates block • Cycle repeats for next -

Driving Restrictions, Goods Transport Germany 2019 Vehicles Concerned Trucks with a Total Permissible Weight of Over 7.5T, As We
Driving Restrictions, Goods Transport Germany 2019 Vehicles concerned trucks with a total permissible weight of over 7.5t, as well as trucks with trailers in case of business-like/commercial or paid transportation of goods including related empty runs. Area throughout the road and motorway network Prohibition Sundays and public holidays from 00h00 to 22h00 Exceptions (applies also to the additional summer driving restrictions) 1. Combined rail/road goods transport from the shipper to the nearest loading railway station or from the nearest designated unloading railway station to the consignee up to a distance of 200km (no limitation on distance during the additional summer restrictions); also combined sea/road goods transport between the place of loading or unloading and a port situated within a radius of 150km maximum (delivery or loading). 2. Deliveries of fresh milk and other dairy produce, fresh meat and its fresh derivatives, fresh fish, live fish and their fresh derivatives, perishable foodstuffs (fruit and vegetables). 3. Transportation of animal by-products according to category 1, Art. 8 as well as category 2, Art. 9f of regulation (EG) No. 1069/2009. 4. Use of vehicles of recovery, towing and breakdown services in case of an accident or other emergencies. 5. Transport of living bees. 6. Empty vehicles, in connection with the transport operations mentioned under point 2-5. 7. Transport operations using vehicles subject to the Federal Law on the obligations of service; the relevant authorisation must be carried on board and produced for inspection on request. Also exempted from the prohibition are vehicles belonging to the police and federal border guard, fire brigades and emergency services, the federal armed services and allied troops. -

Memoriu De Prezentare
PARTENERIAT CU NATURA MEMORIU DE PREZENTARE AUTOSTRADA DE CENTURĂ BUCUREŞTI km 0+000 – km 100+900 COMPANIA NAŢIONALĂ DE ADMINISTRARE A INFRASTRUCTURII RUTIERE SA (CNAIR) CNAIR SA Autostrada de Centură Bucureşti Memoriu de prezentare MEMORIU DE PREZENTARE AUTOSTRADA DE CENTURĂ BUCUREŞTI km 0+000 – km 100+900 Colectiv de elaborare (CE): Ing. Răzvan DUMITRU Ing. Alexandra DOBA Geograf Corneliu BUJOR Biolog Cristina RĂDUCANU Ecolog Ionuţ STAMAT Drd. Geograf Dragoş MĂNTOIU MSc. Ecolog Silvia BORLEA Descrierea documentului şi revizii Verificat Rev. nr. Detalii Data Autor Aprobat de Tehnic Calitate 00 Draft intern Mai 2020 CE AD AD - Memoriu de prezentare – AD AD - 01 Iunie 2020 CE versiune draft 02 Memoriu de prezentare Iunie 2020 CE AD AD MN Referinţă document: Memoriu prezentare_Autostrada de Centura Bucuresti_rev02 Lista de distribuţie Rev Destinatar Copia. nr. Format Confidenţialitate CNAIR 1, 2 Printat, Electronic 02 Alsim Alarko 1 Electronic Da EPC Consultanţă de mediu SRL 1 Electronic Verificat: Aprobat: Ing. Alexandra DOBA (AD) Dr. Ecol. Marius NISTORESCU (MN) Director Tehnic, EPC Director General, EPC 2 CNAIR SA Autostrada de Centură Bucureşti Memoriu de prezentare A nu se copia! 3 CNAIR SA Autostrada de Centură Bucureşti Memoriu de prezentare CUPRINS 1 DENUMIREA PROIECTULUI ........................................................................................................... 8 2 TITULARUL PROIECTULUI ............................................................................................................. 8 3 DESCRIEREA CARACTERISTICILOR -

Paper Was to Explore to Highlight the Major Changes in Human Behavior Using the Analysis of Road Traffic Data As a Representation of Activities and Interaction
Traffic Analysis Studies Using GIS Tools Ana Cornelia BADEA, Gheorghe BADEA, Romania Key words: Geoinformation/GI; Spatial planning; traffic; GIS; real time SUMMARY This article highlights the advantages offered by current GIS technology to perform various traffic analyzes. For this purpose, several areas in Romania that are very crowded at certain times of the week, for example on weekends using were chosen and analyzes using real-time traffic services and historical data for a certain date and time to make comparisons were performed. These traffic analyzes are extremely useful to substantiate the decisions of the authorities for the improvement of the road infrastructure in order to streamline the traffic. REZUMAT Acest articol evidențiază avantajele oferite de tehnologia GIS actuală pentru efectuarea diverselor analize de trafic. În acest scop, au fost alese și analizate câteva zone din România care sunt foarte aglomerate în anumite momente ale săptămânii, de exemplu în weekend, folosind servicii de trafic în timp real și date istorice pentru o anumită dată și oră pentru a face comparații. Aceste analize de trafic sunt extrem de utile pentru a fundamenta deciziile autorităților privind îmbunătățirea infrastructurii rutiere în vederea eficientizării traficului. Traffic Analysis Studies Using GIS Tools (11169) Ana Cornelia Badea and Gheorghe Badea (Romania) FIG e-Working Week 2021 Smart Surveyors for Land and Water Management - Challenges in a New Reality Virtually in the Netherlands, 21–25 June 2021 Traffic Analysis Studies Using GIS Tools Ana Cornelia BADEA, Gheorghe BADEA, Romania 1. INTRODUCTION There are currently a number of geospatial web solutions that provide real-time traffic status and some that include historical data. -

COST Action CA16109 COLOSSAL ̶ Working Groups 2 and 4, and MC Meeting 24-28 Sep 2018 Bucharest, Romania
COST Action CA16109 COLOSSAL ̶ Working Groups 2 and 4, and MC Meeting 24-28 Sep 2018 Bucharest, Romania COST Action CA16109 Chemical On-Line cOmpoSition and Source Apportionment of fine aerosoL COLOSSAL Working Groups 2 and 4 and Management Committee Meeting 24-28 Sep 2018, Bucharest, Romania CONFERENCE LOCATION National Institute for Aerospace Research "Elie Carafoli" - INCAS Bucharest bvd Iuliu Maniu 220 Local organizer: Luminita Marmureanu (INOE), [email protected] INCAS - National Institute for Aerospace Research "Elie Carafoli" is the leading research establishment in aerospace sciences in Romania, with more than 60 years tradition in aerospace engineering, flow physics and applied aerodynamics, using state-of-the-art technologies and unique infrastructure of national strategic importance. Please find below the link with the exact location: http://www.incas.ro/index.php?option=com_contact&view=contact&id=1&Itemid=56 Easiest way to reach INCAS (bvd Iuliu Maniu 220) is by subway, the station is called Pacii and is 1 minute walking. The subway line is M3, 7 stations from “Unirii” station (the center of Bucharest). 1 COST Action CA16109 COLOSSAL ̶ Working Groups 2 and 4, and MC Meeting 24-28 Sep 2018 Bucharest, Romania TRAVEL TO BUCHAREST 1) Geo-politics Romania is a medium-sized European country, with a population of 18 mil. inhabitants, now part of the European Union. The national currency is Romanian LEU (RON): 1 EUR 4.5 RON. Bucharest is the capital of Romania. With an official population of approximately 2 million inhabitants, Bucharest is the sixth largest city in Europe, after London, Paris, Berlin, Madrid and Rome.