Evolution of Green Plants As Deduced from 5S Rrna Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Seed Plant Models
Review Tansley insight Why we need more non-seed plant models Author for correspondence: Stefan A. Rensing1,2 Stefan A. Rensing 1 2 Tel: +49 6421 28 21940 Faculty of Biology, University of Marburg, Karl-von-Frisch-Str. 8, 35043 Marburg, Germany; BIOSS Biological Signalling Studies, Email: stefan.rensing@biologie. University of Freiburg, Sch€anzlestraße 18, 79104 Freiburg, Germany uni-marburg.de Received: 30 October 2016 Accepted: 18 December 2016 Contents Summary 1 V. What do we need? 4 I. Introduction 1 VI. Conclusions 5 II. Evo-devo: inference of how plants evolved 2 Acknowledgements 5 III. We need more diversity 2 References 5 IV. Genomes are necessary, but not sufficient 3 Summary New Phytologist (2017) Out of a hundred sequenced and published land plant genomes, four are not of flowering plants. doi: 10.1111/nph.14464 This severely skewed taxonomic sampling hinders our comprehension of land plant evolution at large. Moreover, most genetically accessible model species are flowering plants as well. If we are Key words: Charophyta, evolution, fern, to gain a deeper understanding of how plants evolved and still evolve, and which of their hornwort, liverwort, moss, Streptophyta. developmental patterns are ancestral or derived, we need to study a more diverse set of plants. Here, I thus argue that we need to sequence genomes of so far neglected lineages, and that we need to develop more non-seed plant model species. revealed much, the exact branching order and evolution of the I. Introduction nonbilaterian lineages is still disputed (Lanna, 2015). Research on animals has for a long time relied on a number of The first (small) plant genome to be sequenced was of THE traditional model organisms, such as mouse, fruit fly, zebrafish or model plant, the weed Arabidopsis thaliana (c. -

Math/C SC 5610 Computational Biology Lecture 12: Phylogenetics
Math/C SC 5610 Computational Biology Lecture 12: Phylogenetics Stephen Billups University of Colorado at Denver Math/C SC 5610Computational Biology – p.1/25 Announcements Project Guidelines and Ideas are posted. (proposal due March 8) CCB Seminar, Friday (Mar. 4) Speaker: Jack Horner, SAIC Title: Phylogenetic Methods for Characterizing the Signature of Stage I Ovarian Cancer in Serum Protein Mas Time: 11-12 (Followed by lunch) Place: Media Center, AU008 Math/C SC 5610Computational Biology – p.2/25 Outline Distance based methods for phylogenetics UPGMA WPGMA Neighbor-Joining Character based methods Maximum Likelihood Maximum Parsimony Math/C SC 5610Computational Biology – p.3/25 Review: Distance Based Clustering Methods Main Idea: Requires a distance matrix D, (defining distances between each pair of elements). Repeatedly group together closest elements. Different algorithms differ by how they treat distances between groups. UPGMA (unweighted pair group method with arithmetic mean). WPGMA (weighted pair group method with arithmetic mean). Math/C SC 5610Computational Biology – p.4/25 UPGMA 1. Initialize C to the n singleton clusters f1g; : : : ; fng. 2. Initialize dist(c; d) on C by defining dist(fig; fjg) = D(i; j): 3. Repeat n ¡ 1 times: (a) determine pair c; d of clusters in C such that dist(c; d) is minimal; define dmin = dist(c; d). (b) define new cluster e = c S d; update C = C ¡ fc; dg Sfeg. (c) define a node with label e and daughters c; d, where e has distance dmin=2 to its leaves. (d) define for all f 2 C with f 6= e, dist(c; f) + dist(d; f) dist(e; f) = dist(f; e) = : (avg. -

Understanding the Processes Underpinning Patterns Of
Opinion Understanding the Processes Underpinning Patterns of Phylogenetic Regionalization 1,2, 3,4 1 Barnabas H. Daru, * Tammy L. Elliott, Daniel S. Park, and 5,6 T. Jonathan Davies A key step in understanding the distribution of biodiversity is the grouping of regions based on their shared elements. Historically, regionalization schemes have been largely species centric. Recently, there has been interest in incor- porating phylogenetic information into regionalization schemes. Phylogenetic regionalization can provide novel insights into the mechanisms that generate, distribute, and maintain biodiversity. We argue that four processes (dispersal limitation, extinction, speciation, and niche conservatism) underlie the forma- tion of species assemblages into phylogenetically distinct biogeographic units. We outline how it can be possible to distinguish among these processes, and identify centers of evolutionary radiation, museums of diversity, and extinction hotspots. We suggest that phylogenetic regionalization provides a rigorous and objective classification of regional diversity and enhances our knowledge of biodiversity patterns. Biogeographical Regionalization in a Phylogenetic Era 1 Department of Organismic and Biogeographical boundaries delineate the basic macrounits of diversity in biogeography, Evolutionary Biology, Harvard conservation, and macroecology. Their location and composition of species on either side of University, Cambridge, MA 02138, these boundaries can reflect the historical processes that have shaped the present-day USA th 2 Department of Plant Sciences, distribution of biodiversity [1]. During the early 19 century, de Candolle [2] created one of the University of Pretoria, Private Bag first global geographic regionalization schemes for plant diversity based on both ecological X20, Hatfield 0028, Pretoria, South and historical information. This was followed by Sclater [3], who defined six zoological regions Africa 3 Department of Biological Sciences, based on the global distribution of birds. -

Evolution of Land Plants P
Chapter 4. The evolutionary classification of land plants The evolutionary classification of land plants Land plants evolved from a group of green algae, possibly as early as 500–600 million years ago. Their closest living relatives in the algal realm are a group of freshwater algae known as stoneworts or Charophyta. According to the fossil record, the charophytes' growth form has changed little since the divergence of lineages, so we know that early land plants evolved from a branched, filamentous alga dwelling in shallow fresh water, perhaps at the edge of seasonally-desiccating pools. The biggest challenge that early land plants had to face ca. 500 million years ago was surviving in dry, non-submerged environments. Algae extract nutrients and light from the water that surrounds them. Those few algae that anchor themselves to the bottom of the waterbody do so to prevent being carried away by currents, but do not extract resources from the underlying substrate. Nutrients such as nitrogen and phosphorus, together with CO2 and sunlight, are all taken by the algae from the surrounding waters. Land plants, in contrast, must extract nutrients from the ground and capture CO2 and sunlight from the atmosphere. The first terrestrial plants were very similar to modern mosses and liverworts, in a group called Bryophytes (from Greek bryos=moss, and phyton=plants; hence “moss-like plants”). They possessed little root-like hairs called rhizoids, which collected nutrients from the ground. Like their algal ancestors, they could not withstand prolonged desiccation and restricted their life cycle to shaded, damp habitats, or, in some cases, evolved the ability to completely dry-out, putting their metabolism on hold and reviving when more water arrived, as in the modern “resurrection plants” (Selaginella). -

A Review of the Late Jurassic–Early Cretaceous Charophytes from The
A review of the Late Jurassic–Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France Roch-Alexandre Benoit, Didier Néraudeau, Carles Martin-Closas To cite this version: Roch-Alexandre Benoit, Didier Néraudeau, Carles Martin-Closas. A review of the Late Jurassic– Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France. Cretaceous Research, Elsevier, 2017, 79, pp.199-213. <10.1016/j.cretres.2017.07.009>. <insu-01574653> HAL Id: insu-01574653 https://hal-insu.archives-ouvertes.fr/insu-01574653 Submitted on 16 Aug 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Accepted Manuscript A review of the Late Jurassic–Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France Roch-Alexandre Benoit, Didier Neraudeau, Carles Martín-Closas PII: S0195-6671(17)30121-0 DOI: 10.1016/j.cretres.2017.07.009 Reference: YCRES 3658 To appear in: Cretaceous Research Received Date: 13 March 2017 Revised Date: 5 July 2017 Accepted Date: 17 July 2017 Please cite this article as: Benoit, R.-A., Neraudeau, D., Martín-Closas, C., A review of the Late Jurassic–Early Cretaceous charophytes from the northern Aquitaine Basin in south-west France, Cretaceous Research (2017), doi: 10.1016/j.cretres.2017.07.009. -

Supplementary Information the Biodiversity and Geochemistry Of
Supplementary Information The Biodiversity and Geochemistry of Cryoconite Holes in Queen Maud Land, East Antarctica Figure S1. Principal component analysis of the bacterial OTUs. Samples cluster according to habitats. Figure S2. Principal component analysis of the eukaryotic OTUs. Samples cluster according to habitats. Figure S3. Principal component analysis of selected trace elements that cause the separation (primarily Zr, Ba and Sr). Figure S4. Partial canonical correspondence analysis of the bacterial abundances and all non-collinear environmental variables (i.e., after identification and exclusion of redundant predictor variables) and without spatial effects. Samples from Lake 3 in Utsteinen clustered with higher nitrate concentration and samples from Dubois with a higher TC abundance. Otherwise no clear trends could be observed. Table S1. Number of sequences before and after quality control for bacterial and eukaryotic sequences, respectively. 16S 18S Sample ID Before quality After quality Before quality After quality filtering filtering filtering filtering PES17_36 79285 71418 112519 112201 PES17_38 115832 111434 44238 44166 PES17_39 128336 123761 31865 31789 PES17_40 107580 104609 27128 27074 PES17_42 225182 218495 103515 103323 PES17_43 219156 213095 67378 67199 PES17_47 82531 79949 60130 59998 PES17_48 123666 120275 64459 64306 PES17_49 163446 158674 126366 126115 PES17_50 107304 104667 158362 158063 PES17_51 95033 93296 - - PES17_52 113682 110463 119486 119205 PES17_53 126238 122760 72656 72461 PES17_54 120805 117807 181725 181281 PES17_55 112134 108809 146821 146408 PES17_56 193142 187986 154063 153724 PES17_59 226518 220298 32560 32444 PES17_60 186567 182136 213031 212325 PES17_61 143702 140104 155784 155222 PES17_62 104661 102291 - - PES17_63 114068 111261 101205 100998 PES17_64 101054 98423 70930 70674 PES17_65 117504 113810 192746 192282 Total 3107426 3015821 2236967 2231258 Table S2. -

The Charophytes (Charophyta) Locality in the Milkha Stream, Lower Jordan, Israel
Natural Resources and Conservation 3(2): 19-30, 2015 http://www.hrpub.org DOI: 10.13189/nrc.2015.030201 The Charophytes (Charophyta) Locality in the Milkha Stream, Lower Jordan, Israel Sophia Barinova1,*, Roman Romanov2 1Institute of Evolution, University of Haifa, Israel 2Central Siberian Botanical Garden of the Siberian Branch of the Russian Academy of Sciences, Russia Copyright © 2015 by authors, all rights reserved. Authors agree that this article remains permanently open access under the terms of the Creative Commons Attribution License 4.0 International License Abstract First study of new locality the Milkha Stream, revealed 14 charophyte species (16 with ifraspecific variety) the Lower Jordan River tributary, with charophyte algae, in that known for Israel [3,4] from references and our studies. semi-arid region of Israel has been implemented for Last year research in Israel and Eastern Mediterranean let us revealing of algal diversity and ecological assessment of the too includes few new localities, algal diversity of which has water object environment by bio-indication methods. been never studied before [5-10]. Altogether forty seven species from five taxonomic The charophytes prefer alkaline water environment which Divisions of algae and cyanobacteria including one of them forms on the carbonates that are very distributed in studied macro-algae Chara gymnophylla A. Braun were revealed in region. This alkaline freshwater environment can be assessed the Milkha stream. Chara was found in growth in the lower as perspective for find new, unstudied aquatic objects in part of studied stream but away from community in followed which can be identified charophyte algae. The most years. -

Molecular Phylogenetics (Hannes Luz)
Tree: minimum but fully connected (no loop, one breaks) Molecular Phylogenetics (Hannes Luz) Contents: • Phylogenetic Trees, basic notions • A character based method: Maximum Parsimony • Trees from distances • Markov Models of Sequence Evolution, Maximum Likelihood Trees References for lectures • Joseph Felsenstein, Inferring Phylogenies, Sinauer Associates (2004) • Dan Graur, Weng-Hsiun Li, Fundamentals of Molecular Evolution, Sinauer Associates • D.W. Mount. Bioinformatics: Sequences and Genome analysis, 2001. • D.L. Swofford, G.J. Olsen, P.J.Waddell & D.M. Hillis, Phylogenetic Inference, in: D.M. Hillis (ed.), Molecular Systematics, 2 ed., Sunder- land Mass., 1996. • R. Durbin, S. Eddy, A. Krogh & G. Mitchison, Biological sequence analysis, Cambridge, 1998 References for lectures, cont’d • S. Rahmann, Spezielle Methoden und Anwendungen der Statistik in der Bioinformatik (http://www.molgen.mpg.de/~rahmann/afw-rahmann. pdf) • K. Schmid, A Phylogenetic Parsimony Method Considering Neigh- bored Gaps (Bachelor thesis, FU Berlin, 2007) • Martin Vingron, Hannes Luz, Jens Stoye, Lecture notes on ’Al- gorithms for Phylogenetic Reconstructions’, http://lectures.molgen. mpg.de/Algorithmische_Bioinformatik_WS0405/phylogeny_script.pdf Recommended reading/watching • Video streams of Arndt von Haeseler’s lectures held at the Otto Warburg Summer School on Evolutionary Genomics 2006 (http: //ows.molgen.mpg.de/2006/von_haeseler.shtml) • Dirk Metzler, Algorithmen und Modelle der Bioinformatik, http://www. cs.uni-frankfurt.de/~metzler/WS0708/skriptWS0708.pdf Software links • Felsenstein’s list of software packages: http://evolution.genetics.washington.edu/phylip/software.html • PHYLIP is Felsenstein’s free software package for inferring phyloge- nies, http://evolution.genetics.washington.edu/phylip.html • Webinterface for PHYLIP maintained at Institute Pasteur, http://bioweb.pasteur.fr/seqanal/phylogeny/phylip-uk.html • Puzzle (Strimmer, v. -

Conifers Exhibit a Characteristic Inactivation of Auxin to Maintain Tissue
bioRxiv preprint doi: https://doi.org/10.1101/789420; this version posted October 1, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Title Conifers exhibit a characteristic inactivation of auxin to maintain tissue homeostasis Authors Federica Brunoni1,2,a, Silvio Collani1, Rubén Casanova-Saéz2, Jan Šimura2, Michal Karady2,a, Markus Schmid1, Karin Ljung2,b and Catherine Bellini1,3,b 1 Umeå Plant Science Centre, Dept of Plant Physiology, Umeå University (Umu), Umeå, Sweden 2 Umeå Plant Science Centre, Dept of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences (SLU), Umeå, Sweden 3 Institut Jean-Pierre Bourgin, UMR1318 INRA-AgroParisTech Versailles, France a Present address: Laboratory of Growth Regulators, Faculty of Science, Palacký University & Institute of Experimental Botany, The Czech Academy of Sciences, Šlechtitelů 27, CZ- 78371, Olomouc, Czech Republic b to whom correspondence may be addressed. Email: [email protected] or [email protected] or [email protected] Summary • Dynamic regulation of the levels of the natural auxin, indol-3-acetic acid (IAA), is essential to coordinate most of the physiological and developmental processes and responses to environmental changes. Oxidation of IAA is a major pathway to control auxin concentrations in Arabidopsis and, along with IAA conjugation, to respond to perturbation of IAA homeostasis. However, these regulatory mechanisms are still poorly investigated in conifers. To reduce this gap of knowledge, we investigated the different contribution of the IAA inactivation pathways in conifers. • Mass spectrometry-based quantification of IAA metabolites under steady state conditions and after perturbation was investigated to evaluate IAA homeostasis in bioRxiv preprint doi: https://doi.org/10.1101/789420; this version posted October 1, 2019. -

5 Computational Methods and Tools Introductory Remarks by the Chapter Editor, Joris Van Zundert
5 Computational methods and tools Introductory remarks by the chapter editor, Joris van Zundert This chapter may well be the hardest in the book for those that are not all that computationally, mathematically, or especially graph-theoretically inclined. Textual scholars often take to text almost naturally but have a harder time grasping, let alone liking, mathematics. A scholar of history or texts may well go through decades of a career without encountering any maths beyond the basic schooling in arith- metic, algebra, and probability calculation that comes with general education. But, as digital techniques and computational methods progressed and developed, it tran- spired that this field of maths and digital computation had some bearing on textual scholarship too. Armin Hoenen, in section 5.1, introduces us to the early history of computational stemmatology, depicting its early beginnings in the 1950s and point- ing out some even earlier roots. The strong influence of phylogenetics and bioinfor- matics in the 1990s is recounted, and their most important concepts are introduced. At the same time, Hoenen warns us of the potential misunderstandings that may arise from the influx of these new methods into stemmatology. The historical over- view ends with current and new developments, among them the creation of artificial traditions for validation purposes, which is actually a venture with surprisingly old roots. Hoenen’s history shows how a branch of computational stemmatics was added to the field of textual scholarship. Basically, both textual and phylogenetic theory showed that computation could be applied to the problems of genealogy of both textual traditions and biological evolution. -
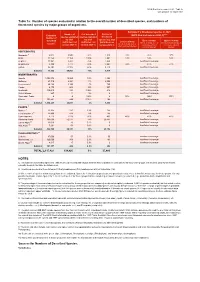
2021-1 RL Stats Table1a.Pdf
IUCN Red List version 2021-1: Table 1a Last updated: 25 March 2021 Table 1a: Number of species evaluated in relation to the overall number of described species, and numbers of threatened species by major groups of organisms. Estimated % threatened species in 2021 Number of % of described Number of 2,3,4 Estimated (IUCN Red List version 2021-1) species evaluated species evaluated threatened Number of 2 by 2021 by 2021 species by 2021 Best estimate Upper estimate described Lower estimate 1 (IUCN Red List (IUCN Red List (IUCN Red List (threatened spp. as % of (threatened and DD spp. as species (threatened spp. as % of version 2021-1) version 2021-1) extant data sufficient % of extant evaluated version 2021-1) extant evaluated species) evaluated species) species) VERTEBRATES Mammals 5 6,513 5,940 91% 1,323 23% 26% 37% Birds 11,158 11,158 100% 1,481 13% 14% 14% Reptiles 11,341 8,492 75% 1,458 Insufficient coverage Amphibians 8,309 7,212 87% 2,442 34% 41% 51% Fishes 35,797 22,005 61% 3,210 Insufficient coverage Subtotal 73,118 54,807 75% 9,914 INVERTEBRATES Insects 1,053,578 10,865 1.0% 1,926 Insufficient coverage Molluscs 81,719 8,881 11% 2,305 Insufficient coverage Crustaceans6 80,122 3,189 4% 743 Insufficient coverage Corals 2,175 864 40% 237 Insufficient coverage Arachnids 110,615 393 0.36% 218 Insufficient coverage Velvet Worms 227 11 5% 9 Insufficient coverage Horseshoe Crabs 4 4 100% 2 50% 100% 100% Others 151,801 844 0.56% 148 Insufficient coverage Subtotal 1,480,241 25,051 2% 5,588 PLANTS 7 Mosses 8 21,925 282 1.3% 165 Insufficient coverage Ferns and Allies 9 11,800 678 6% 266 Insufficient coverage Gymnosperms 1,113 1,016 91% 403 40% 41% 42% Flowering Plants 369,000 52,077 14% 20,883 Insufficient coverage Green Algae 10 11,616 16 0.1% 0 Insufficient coverage Red Algae 10 7,291 58 0.8% 9 Insufficient coverage Subtotal 422,745 54,127 13% 21,726 FUNGI & PROTISTS 11 Lichens 17,000 57 0.3% 50 Insufficient coverage Mushrooms, etc. -

The Anâtaxis Phylogenetic Reconstruction Algorithm
U´ G` F´ Departement´ d’informatique Professeur Bastien Chopard Institut Suisse de Bioinformatique Dr Gabriel Bittar The Anataxisˆ phylogenetic reconstruction algorithm THESE` present´ ee´ a` la Faculte´ des sciences de l’Universite´ de Geneve` pour obtenir le grade de Docteur es` sciences, mention bioinformatique par Bernhard Pascal Sonderegger de Heiden (AR) These` No 3863 Geneve` Atelier d’impression de la Section de Physique 2007 FACULTE´ DES SCIENCES Doctorat es` Sciences mention bioinformatique These` de Monsieur Bernhard SONDEREGGER Intitulee:´ The Anataxisˆ phylogenetic reconstruction algorithm La faculte´ des sciences, sur le preavis´ de Messieurs B. CHOPARD, professeur ad- joint et directeur de these` (Departement´ d’ informatique), G. Bittar, Docteur et co- directeur de these` (Institut Suisse de Bioinformatique, Geneve,` Suisse), A. BAIROCH, professeur adjoint (Faculte´ de medecine,´ Section de medecine´ fon- dementale, Departement´ de biologie structurale et bioinformatique) et N. SALAMIN, docteur (Universite´ de Lausanne, Faculte´ de biologie et de mede- cine Departement´ d’ecologie´ et evolution,´ Lausanne, Suisse), autorise l’impression de la presente´ these,` sans exprimer d’opinion sur les propositions qui y sont enonc´ ees.´ Geneve,` le 26.06.2007 Th`ese-3863- Le Doyen, Pierre SPIERER Contents Contents i Remerciements 1 Preface 1 R´esum´een franc¸ais 5 Introduction a` la phylogen´ etique´ ....................... 5 L’algorithme Anataxisˆ ............................. 8 Calcul de dissimilitudes ............................ 11 Validation numerique´ .............................. 11 Implementation´ ................................. 12 Exemple biologique ............................... 13 Conclusion .................................... 14 1 An introduction to phylogenetics 17 1.1 Homology and homoplasy ........................ 18 1.1.1 Characters and their states .................... 18 1.1.2 Homology is a phylogenetic hypothesis ............ 20 1.1.3 Homoplasy, a pitfall in phylogenetics ............