Reclassification of Pasteurella Gallinarum, [Haemophilus]
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diagnosis of Gallibacterium Anatis in Layers: First Report in Turkey
Brazilian Journal of Poultry Science Revista Brasileira de Ciência Avícola ISSN 1516-635X 2019 / v.21 / n.3 / 001-008 Diagnosis of Gallibacterium Anatis in Layers: First Report in Turkey http://dx.doi.org/10.1590/1806-9061-2019-1019 Original Article Author(s) ABSTRACT Yaman SI https://orcid.org/0000-0002-9998-3806 Gallibacterium anatis, a member of the Pasteurellaceae family, leads Sahan Yapicier OII https://orcid.org/0000-0003-3579-9425 to decrease in egg-production, animal welfare and increase in mortality. I Burdur Mehmet Akif Ersoy University, The Health This study aimed to diagnose G. Anatis, which caused economic losses Sciences Institute, Department of Microbiology, in laying hens by using conventional and molecular techniques. In 15030, Burdur, Turkey. II BurdurMehmet Akif Ersoy University, Faculty of this study, G. anatis was examined from a total of 200 dead chicken Veterinary Medicine, Department of Microbiology, tissues (heart, liver, lung, spleen and trachea) in laying hen farms 15030, Burdur, Turkey. that observed a decrease in egg production with respiratory system infection. Conventional methods based on colony morphology, sugar fermentation tests and hemolytic properties and molecular conformation using 16S rRNA-23S rRNA specific primers were performed to identify G. anatis. G. anatis was isolated in 20 (10%) of the examined samples and isolates were confirmed by conventional PCR. A total of 11 (2.2%) positivity was obtained as isolates were the result of PCR performed on tissues and organs directly. As a result, the presence of G. anatis was detected for the first time in Turkey by this study. It was thought that G. -

Bacterial Communities of the Upper Respiratory Tract of Turkeys
www.nature.com/scientificreports OPEN Bacterial communities of the upper respiratory tract of turkeys Olimpia Kursa1*, Grzegorz Tomczyk1, Anna Sawicka‑Durkalec1, Aleksandra Giza2 & Magdalena Słomiany‑Szwarc2 The respiratory tracts of turkeys play important roles in the overall health and performance of the birds. Understanding the bacterial communities present in the respiratory tracts of turkeys can be helpful to better understand the interactions between commensal or symbiotic microorganisms and other pathogenic bacteria or viral infections. The aim of this study was the characterization of the bacterial communities of upper respiratory tracks in commercial turkeys using NGS sequencing by the amplifcation of 16S rRNA gene with primers designed for hypervariable regions V3 and V4 (MiSeq, Illumina). From 10 phyla identifed in upper respiratory tract in turkeys, the most dominated phyla were Firmicutes and Proteobacteria. Diferences in composition of bacterial diversity were found at the family and genus level. At the genus level, the turkey sequences present in respiratory tract represent 144 established bacteria. Several respiratory pathogens that contribute to the development of infections in the respiratory system of birds were identifed, including the presence of Ornithobacterium and Mycoplasma OTUs. These results obtained in this study supply information about bacterial composition and diversity of the turkey upper respiratory tract. Knowledge about bacteria present in the respiratory tract and the roles they can play in infections can be useful in controlling, diagnosing and treating commercial turkey focks. Next-generation sequencing has resulted in a marked increase in culture-independent studies characterizing the microbiome of humans and animals1–6. Much of these works have been focused on the gut microbiome of humans and other production animals 7–11. -

Identification of Pasteurella Species and Morphologically Similar Organisms
UK Standards for Microbiology Investigations Identification of Pasteurella species and Morphologically Similar Organisms Issued by the Standards Unit, Microbiology Services, PHE Bacteriology – Identification | ID 13 | Issue no: 3 | Issue date: 04.02.15 | Page: 1 of 28 © Crown copyright 2015 Identification of Pasteurella species and Morphologically Similar Organisms Acknowledgments UK Standards for Microbiology Investigations (SMIs) are developed under the auspices of Public Health England (PHE) working in partnership with the National Health Service (NHS), Public Health Wales and with the professional organisations whose logos are displayed below and listed on the website https://www.gov.uk/uk- standards-for-microbiology-investigations-smi-quality-and-consistency-in-clinical- laboratories. SMIs are developed, reviewed and revised by various working groups which are overseen by a steering committee (see https://www.gov.uk/government/groups/standards-for-microbiology-investigations- steering-committee). The contributions of many individuals in clinical, specialist and reference laboratories who have provided information and comments during the development of this document are acknowledged. We are grateful to the Medical Editors for editing the medical content. For further information please contact us at: Standards Unit Microbiology Services Public Health England 61 Colindale Avenue London NW9 5EQ E-mail: [email protected] Website: https://www.gov.uk/uk-standards-for-microbiology-investigations-smi-quality- and-consistency-in-clinical-laboratories UK Standards for Microbiology Investigations are produced in association with: Logos correct at time of publishing. Bacteriology – Identification | ID 13 | Issue no: 3 | Issue date: 04.02.15 | Page: 2 of 28 UK Standards for Microbiology Investigations | Issued by the Standards Unit, Public Health England Identification of Pasteurella species and Morphologically Similar Organisms Contents ACKNOWLEDGMENTS ......................................................................................................... -

Phenotypic and Molecular Characterization of the Capsular Serotypes of Pasteurella Multocida Isolates from Pneumonic Cases of Cattle in Ethiopia
Phenotypic and Molecular Characterization of the Capsular Serotypes of Pasteurella multocida Isolates from Pneumonic Cases of Cattle in Ethiopia Mirtneh Akalu Yilma ( [email protected] ) Koneru Lakshmaiah Education Foundation https://orcid.org/0000-0001-5936-6873 Murthy Bhadra Vemulapati Koneru Lakshmaiah Education Foundation Takele Abayneh Tefera Veterinaerinstituttet Martha Yami VeterinaryInstitute Teferi Degefa Negi VeterinaryInstitue Alebachew Belay VeterinaryInstitute Getaw Derese VeterinaryInstitute Esayas Gelaye Leykun Veterinaerinstituttet Research article Keywords: Biovar, Capsular type, Cattle, Ethiopia, Pasteurella multocida Posted Date: January 19th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-61749/v2 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/13 Abstract Background: Pasteurella multocida is a heterogeneous species and opportunistic pathogen associated with pneumonia in cattle. Losses due to pneumonia and associated expenses are estimated to be higher in Ethiopia with limited information about the distribution of capsular serotypes. Hence, this study was designed to determine the phenotypic and capsular serotypes of P. multocida from pneumonic cases of cattle. Methods: A cross sectional study with purposive sampling method was employed in 400 cattle from April 2018 to January 2019. Nasopharyngeal swabs and lung tissue samples were collected from clinically suspected pneumonic cases of calves (n = 170) and adult cattle (n = 230). Samples were analyzed using bacteriological and molecular assay. Results: Bacteriological analysis revealed isolation of 61 (15.25%) P. multocida subspecies multocida. Incidence was higher in calves 35 (57.38%) compared to adult cattle 26 (42.62%) at P < 0.5. PCR assay targeting KMT1 gene (~460 bp) conrmed P. -

Gallibacterium Anatis: an Emerging Pathogen of Poultry Birds And
ary Scien in ce r te & e T V e f c h o Journal of Veterinary Science & n n l o o a a l l n n o o r r g g u u Singh, et al., J Veterinar Sci Techno 2016, 7:3 y y o o J J Technology DOI: 10.4172/2157-7579.1000324 ISSN: 2157-7579 Review Article Open Access Gallibacterium anatis: An Emerging Pathogen of Poultry Birds and Domiciled Birds Shiv Varan Singh, Bhoj R Singh*, Dharmendra K Sinha, Vinodh Kumar OR, Prasanna Vadhana A, Monika Bhardwaj and Sakshi Dubey Division of Epidemiology, ICAR-Indian Veterinary Research Institute, Izatnagar-243 122, Uttar Pradesh, India *Corresponding author: Dr. Bhoj R Singh, Acting Head of Division of Epidemiology, ICAR-IVRI, Izatnagar-243122, Uttar Pradesh, India, Tel: +91-8449033222; E-mail: [email protected] Rec date: Feb 09, 2016; Acc date: Mar 16, 2016; Pub date: Mar 18, 2016 Copyright: © 2016 Singh SV, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Gallibacterium anatis though known since long as opportunistic pathogen of intensively reared poultry birds has emerged in last few years as multiple drug resistance pathogen causing heavy mortality outbreaks not only in poultry birds but also in other domiciled or domestic birds. Due to its fastidious nature, commensal status and with no pathgnomonic lesions in diseased birds G. anatis infection often remains obscure for diagnosis. -

Pasteurellaceae: P. Multocida, Avibacterium Gallinarum, A
Pasteurellaceae: P. Multocida, Avibacterium gallinarum, A. paragallinarum All the members of the family Pasteurellaceae are gram negative coccobacilli. They are facultative anaerobes, and typically oxidase-positive (which sets them apart from members of the family Enterobacteriaceae). Morphology and Staining Members of the genera Avibacterium, and Pasteurella are gram-negative coccobacilli. Bipolarity, that is, the staining of only the tips of cells, may be demonstrable with polychrome stains (e.g., Wright’s stain). Cell structure Adhesins. Some and probably all members of the family Pasteurellaceae produce adhesins (and possibly more than one kind). A type 4 fimbria (adhesin) has been described for avian strains of P. Multocida. Capsule. The hyaluronic acid capsule of type A strains P. multocida serves as an adhesin. The hyaluronic acid is similar (if not identical) to host tissue components, and is thus poorly antigenic; they also bind complement components poorly (and is therefore antiphagocytic).The hyaluronic acid capsule also serves as an adhesin for respiratory tract epithelial cells as in the case of capsule type A strains of P. Multocida Exotoxin. Pasteurella produce a number of proteins with toxic activity. At least two of these are important in the pathogenesis of disease: RTX and Rho toxin Growth Characteristics Avibacterium and Pasteurella grow best in the presence of serum or blood. After overnight incubation (35–37 ◦C), colonies are up to 2 mm in diameter, clear to 1 grayish, and smooth or mucoid. All are gram-negative, nonmotile coccobacilli. They are facultative anaerobes, typically oxidase-positive. Variability P. multocida consists of 5 capsular serogroups (A, B, D, E, and F) and 16 somatic serotypes (1–16). -
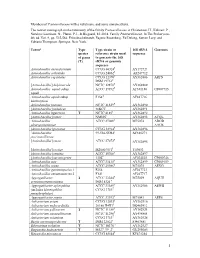
Members of Pasteurellaceae with a Valid Name and Some Unnamed Taxa
Members of Pasteurellaceae with a valid name and some unnamed taxa. The newest monograph on the taxonomy of the family Pasteurellaceae is Christensen, H., Kuhnert, P., Nørskov-Lauritsen, N., Planet, P.J., & Bisgaard, M. 2014. Family Pasteurellaceae. In The Prokaryotes 4th ed. Vol. 9, pp. 535-564. Erko Stackebrandt, Eugene Rosenberg, Ed Delong, Steven Lory, and Fabiano Thompson. Springer, New York. Taxona Type Type strain or 16S rRNA Genomes species reference strain used sequence of genus to generate the 16S (T) rRNA or genomic sequence Actinobacillus anseriformium CCUG 60324T AY172727 Actinobacillus arthritidis CCUG 24862T AF247712 Actinobacillus capsulatus CCUG 12396T AY362886 ARFN DSM 19761T [Actinobacillus] delphinicola NCTC 12870T AY362889 Actinobacillus equuli subsp. ATCC 19392T AF381186 CP007715 equuli Actinobacillus equuli subsp. F154T AF247716 haemolyticus Actinobacillus hominis NCTC 11529T AY362890 [Actinobacillus] indolicus 46KC2T AY362891 Actinobacillus lignieresii T NCTC 4189T AY362892 [Actinobacillus] minor NM305T AY362893 ACQL Actinobacillus ATCC 27088T M75074 ADOD pleuropneumoniae AACK [Actinobacillus] porcinus CCUG 38924T AY362896 ‘Actinobacillus 99-536-55H-F AF486274 porcitonsillarum’ T [Actinobacillus] rossii ATCC 27072 AY362895 [Actinobacillus] scotiae M2000/95/1T Y09653 [Actinobacillus] seminis ATCC 15768T AY362897 [Actinobacillus] succinogenes 130ZT AF024525 CP000746 Actinobacillus suis ATCC 33415T AY362899 CP009159 Actinobacillus ureae ATCC 25986T M75075 AEVG Actinobacillus genomospecies 1 F264 AF247723 Actinobacillus -

Phenotypic and Molecular Characterization of the Capsular Serotypes of Pasteurella Multocida Isolates from Bovine Respiratory Disease Cases in Ethiopia
Phenotypic and Molecular Characterization of the Capsular Serotypes of Pasteurella Multocida Isolates From Bovine Respiratory Disease Cases in Ethiopia Mirtneh Akalu Yilma ( [email protected] ) Koneru Lakshmaiah Education Foundation https://orcid.org/0000-0001-5936-6873 Murthy Bhadra Vemulapati Koneru Lakshmaiah Education Foundation Takele Abayneh Tefera Veterinaerinstituttet Martha Yami VeterinaryInstitute Teferi Degefa Negi VeterinaryInstitue Alebachew Belay VeterinaryInstitute Getaw Derese VeterinaryInstitute Esayas Gelaye Leykun Veterinaerinstituttet Research article Keywords: Antibiogram, Biovar, Capsular type, Cattle, Ethiopia, Pasteurella multocida Posted Date: September 9th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-61749/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/15 Abstract Background: Pasteurella multocida is a heterogeneous species and opportunistic pathogen that causes bovine respiratory disease. This disease is one of an economically important disease in Ethiopia. Losses due to mortality and associated expenses are estimated to be higher in the country. Studies revealed that limited information is available regarding the capsular types, genotypes, and antimicrobial sensitivity of P. multocida isolates circulating in the country. This suggests, further molecular advances to understand the etiological diversity of the pathogens representing severe threats to the cattle population. Results: Bacteriological analysis of nasopharyngeal swab and pneumonic lung tissue samples collected from a total of 400 samples revealed isolation of 61 (15.25%) P. multocida subspecies multocida. 35 (20.59%) were isolated from calves and 26 (11.30%) from adult cattle. Molecular analysis using PCR assay targeting KMT1 gene (~460 bp) amplication was shown in all presumptive isolates. Capsular typing also conrmed the presence of serogroup A (hyaD-hyaC) gene (~1044 bp) and serogroup D (dcbF) gene (~657 bp) from 56 (91.80%) and 5 (8.20%) isolates, respectively. -

Genome Analysis and Phylogenetic Relatedness of Gallibacterium Anatis Strains from Poultry Timothy J
Veterinary Microbiology and Preventive Medicine Veterinary Microbiology and Preventive Medicine Publications 1-24-2013 Genome Analysis and Phylogenetic Relatedness of Gallibacterium anatis Strains from Poultry Timothy J. Johnson University of Minnesota Jessica L. Danzeisen University of Minnesota Darrell W. Trampel Iowa State University, [email protected] Lisa K. Nolan Iowa State University, [email protected] Torsten Seemann FMoolnloaswh U thinivser asitndy additional works at: http://lib.dr.iastate.edu/vmpm_pubs See nePxat pratge of for the addiGetionnomical authors Commons, Large or Food Animal and Equine Medicine Commons, Veterinary Microbiology and Immunobiology Commons, and the Veterinary Preventive Medicine, Epidemiology, and Public Health Commons The ompc lete bibliographic information for this item can be found at http://lib.dr.iastate.edu/ vmpm_pubs/2. For information on how to cite this item, please visit http://lib.dr.iastate.edu/ howtocite.html. This Article is brought to you for free and open access by the Veterinary Microbiology and Preventive Medicine at Iowa State University Digital Repository. It has been accepted for inclusion in Veterinary Microbiology and Preventive Medicine Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Genome Analysis and Phylogenetic Relatedness of Gallibacterium anatis Strains from Poultry Abstract Peritonitis is the major disease problem of laying hens in commercial table egg and parent stock operations. Despite its importance, the etiology and pathogenesis of this disease have not been completely clarified. Although avian pathogenic Escherichia coli (APEC) isolates have been incriminated as the causative agent of laying hen peritonitis, Gallibacterium anatis are frequently isolated from peritonitis lesions. -

Bacterioplankton in the Oregon Upwelling System: Distribution, Cell
AN ABSTRACT OF THE DISSERTATION OF Krista Longnecker for the degree of Doctor ofPhilosophy in Oceanography presented on July 2, 2004. Title: Bacterioplankton in the Oregon UpwellingSystem: Distribution, Cell-specific Leucine Incorporation, and Diversity Abstract approved: Redacted for privacy Barry F. Sherr Marine bacterioplankton playan important role in global elemental cycles because they return carbon dioxide and nutrientsto the biosphere as they reduce organic matter. Furthermore, marine bacterioplanktonare not unifonnly active, and subpopulations of the in situ communitymay be more or less active at any given time. Defining whether or not a cell is 'active' isnot without difficulty, and the result varies depending on the assay used, since differentassays examine different physiological processes within a cell. Linking the level of activity ofa cell with its phylogenetic identity is an additional important step inexamination of the role of marine prokaryotes in global elemental cycles. In this project,flow cytometry was used in two ways to examine relative cell-specific metabolic activity inbacterioplankton cells: as relative cell-specific nucleic acidcontent via staining with SYBR Green I, andas ability to reduce sufficient 5-cyano-2,3-ditolyltetrazolium chloride (CTC) to be identified as having an active electrontransport system. Based on flow cytometric sorting of cells labeled with 3H-leucine, thehigh nucleic acid (HNA) cells had higher cell-specific leucine incorporation rates than thelow nucleic acid (LNA) cells. The HNA cells were also responsible for proportionatelymore of the leucine incorporation by the total heterotrophic population. Whilethe CTC-positive cells had higheraverage cell-specific leucine incorporation rates than theHNA cells, their low abundances meant that they were responsible for less than 15% ofthe total leucine incorporation. -

A Case of Lower Respiratory Tract Infection with Canine-Associated
DOI: 10.7860/JCDR/2015/13900.6351 Case Report A Case of Lower Respiratory Tract Infection with Canine-associated Microbiology Section Microbiology Pasteurella canis in a Patient with Chronic Obstructive Pulmonary Disease SEVITHA BHAT1, PREETAM R. ACHARYA2, DHANASHREE BIRANTHABAIL3, ASEEM RANGNEKAR4, SACHIN SHIRAGAVI5 ABSTRACT This is the report of lower respiratory tract infection with Pasteurella canis in a chronic obstructive pulmonary disease (COPD) patient with history of casual exposure to cats. Pasteurella species are part of the oral and gastrointestinal flora in the canine animals. These organisms are usually implicated in wound infection following animal bites, but can also be associated with a variety of infections including respiratory tract infections. Keywords: Canine animals, Doxycycline, Vitek 2 system CASE REPORT A 70-year-old male, hotel employee by occupation, known case of Chronic obstructive pulmonary disease (COPD) and ischaemic heart disease (IHD) presented to our hospital with a history of cough with purulent expectoration, low grade fever and worsening breathlessness of seven days duration. Patient had history of recurrent exacerbations of COPD caused by Pseudomonas spp. six months back. Patient was an active smoker and gave a history of casual exposure to domestic cats. [Table/Fig-1]: Chest radiograph PA view showing hyper-inflated lung fields and an On examination, patient was conscious, afebrile, tachypneic (res- unfolded aorta [Table/Fig-2]: Culture on Chocolate agar plate showing smooth grey colonies of P.canis piratory rate of 22/minute), mildly hypoxic (oxygen saturation on room air of 88% by pulse oximetry) and haemodynamically stable. Respiratory system examination revealed a barrel shaped chest and bilaterally diminished breath sounds with diffused polyphonic wheeze on auscultation. -

Specific Identification of by a PCR Using Primers Targeting the 16S
Specific identification of by a PCR using primers targeting the 16S rRNA and 23S rRNA genes Anders Miki Bojesen, Maria Elena Vazquez, Fransisco Robles, Carlos Gonzalez, Edgardo V. Soriano, John Elmerdahl Olsen, Henrik Christensen To cite this version: Anders Miki Bojesen, Maria Elena Vazquez, Fransisco Robles, Carlos Gonzalez, Edgardo V. Soriano, et al.. Specific identification of by a PCR using primers targeting the 16S rRNA and23SrRNA genes. Veterinary Microbiology, Elsevier, 2007, 123 (1-3), pp.262. 10.1016/j.vetmic.2007.02.013. hal-00532210 HAL Id: hal-00532210 https://hal.archives-ouvertes.fr/hal-00532210 Submitted on 4 Nov 2010 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Accepted Manuscript Title: Specific identification of Gallibacterium by a PCR using primers targeting the 16S rRNA and 23S rRNA genes Authors: Anders Miki Bojesen, Maria Elena Vazquez, Fransisco Robles, Carlos Gonzalez, Edgardo V. Soriano, John Elmerdahl Olsen, Henrik Christensen PII: S0378-1135(07)00081-8 DOI: doi:10.1016/j.vetmic.2007.02.013 Reference: VETMIC 3598 To appear in: VETMIC Received date: 16-1-2007 Revised date: 7-2-2007 Accepted date: 9-2-2007 Please cite this article as: Bojesen, A.M., Vazquez, M.E., Robles, F., Gonzalez, C., Soriano, E.V., Olsen, J.E., Christensen, H., Specific identification of Gallibacterium by a PCR using primers targeting the 16S rRNA and 23S rRNA genes, Veterinary Microbiology (2007), doi:10.1016/j.vetmic.2007.02.013 This is a PDF file of an unedited manuscript that has been accepted for publication.