Article & Appendix
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Queensland Public Boat Ramps
Queensland public boat ramps Ramp Location Ramp Location Atherton shire Brisbane city (cont.) Tinaroo (Church Street) Tinaroo Falls Dam Shorncliffe (Jetty Street) Cabbage Tree Creek Boat Harbour—north bank Balonne shire Shorncliffe (Sinbad Street) Cabbage Tree Creek Boat Harbour—north bank St George (Bowen Street) Jack Taylor Weir Shorncliffe (Yundah Street) Cabbage Tree Creek Boat Harbour—north bank Banana shire Wynnum (Glenora Street) Wynnum Creek—north bank Baralaba Weir Dawson River Broadsound shire Callide Dam Biloela—Calvale Road (lower ramp) Carmilla Beach (Carmilla Creek Road) Carmilla Creek—south bank, mouth of creek Callide Dam Biloela—Calvale Road (upper ramp) Clairview Beach (Colonial Drive) Clairview Beach Moura Dawson River—8 km west of Moura St Lawrence (Howards Road– Waverley Creek) Bund Creek—north bank Lake Victoria Callide Creek Bundaberg city Theodore Dawson River Bundaberg (Kirby’s Wall) Burnett River—south bank (5 km east of Bundaberg) Beaudesert shire Bundaberg (Queen Street) Burnett River—north bank (downstream) Logan River (Henderson Street– Henderson Reserve) Logan Reserve Bundaberg (Queen Street) Burnett River—north bank (upstream) Biggenden shire Burdekin shire Paradise Dam–Main Dam 500 m upstream from visitors centre Barramundi Creek (Morris Creek Road) via Hodel Road Boonah shire Cromarty Creek (Boat Ramp Road) via Giru (off the Haughton River) Groper Creek settlement Maroon Dam HG Slatter Park (Hinkson Esplanade) downstream from jetty Moogerah Dam AG Muller Park Groper Creek settlement Bowen shire (Hinkson -

Your Labor Member in the Queensland Parliament
YOUR LABOR MEMBER IN THE QUEENSLAND PARLIAMENT Cape Vork / Douglas / Cooktown / Mareeba: Torres Strait / NPA P: 1800816264 - Fax: 07 40312437 P: 1800802391 - Fax: 07 4069 1620 PO Box 2080, Cairns 4870 PO Box 437, Thursday Island 4875 E: [email protected] E: cook.th [email protected] Submission to the Queensland Competition Authority Level 19 12 Creek Street BRISBANE QLD 4001 REVIEW OF SUN WATER PRICING STRUCTURE Submission to the Queensland Competition Authority by Jason O'Brien MP Member for Cook, on behalf of a constituent who is a Sun Water user, requesting changes to the pricing policy of Sun Water to their customers holding pension concession cards who access sun water for their domestic use only. Under the present Sun Water tariff structure they are able to negotiate how much revenue could be collected from variable and fixed charges. My submission is that Sun Water should be free to offer a Pensioner Water Subsidy Scheme for eligible pensions to reduce the impact of increased water price increases. This concession could be in the form of a rebate similar to the rate rebate scheme which applies across the state off local government rates charges. Sun Water must have the ability to provide water services to the community at a reasonable cost taking into account the most effective way to utilise the resource for the community's benefit. To this end pensions accessing Sun Water's resource purely for domestic use should be considered in a wider public interest context. Prices should be cost reflective and take into account relevant public interest matters such as pensioners accessing their resource In conclusion I submit that Sun Water should be providing a Pensioner Water Subsidy Scheme and the Queensland Competition Authority should recognise this as a public interest issue. -

The Non-Human Reservoirs of Ross River Virus: a Systematic Review of the Evidence Eloise B
Stephenson et al. Parasites & Vectors (2018) 11:188 https://doi.org/10.1186/s13071-018-2733-8 REVIEW Open Access The non-human reservoirs of Ross River virus: a systematic review of the evidence Eloise B. Stephenson1*, Alison J. Peel1, Simon A. Reid2, Cassie C. Jansen3,4 and Hamish McCallum1 Abstract: Understanding the non-human reservoirs of zoonotic pathogens is critical for effective disease control, but identifying the relative contributions of the various reservoirs of multi-host pathogens is challenging. For Ross River virus (RRV), knowledge of the transmission dynamics, in particular the role of non-human species, is important. In Australia, RRV accounts for the highest number of human mosquito-borne virus infections. The long held dogma that marsupials are better reservoirs than placental mammals, which are better reservoirs than birds, deserves critical review. We present a review of 50 years of evidence on non-human reservoirs of RRV, which includes experimental infection studies, virus isolation studies and serosurveys. We find that whilst marsupials are competent reservoirs of RRV, there is potential for placental mammals and birds to contribute to transmission dynamics. However, the role of these animals as reservoirs of RRV remains unclear due to fragmented evidence and sampling bias. Future investigations of RRV reservoirs should focus on quantifying complex transmission dynamics across environments. Keywords: Amplifier, Experimental infection, Serology, Virus isolation, Host, Vector-borne disease, Arbovirus Background transmission dynamics among arboviruses has resulted in Vertebrate reservoir hosts multiple definitions for the key term “reservoir” [9]. Given Globally, most pathogens of medical and veterinary im- the diversity of virus-vector-vertebrate host interactions, portance can infect multiple host species [1]. -

Demographic Consequences of Superabundance in Krefft's River
i The comparative ecology of Krefft’s River Turtle Emydura krefftii in Tropical North Queensland. By Dane F. Trembath B.Sc. (Zoology) Applied Ecology Research Group University of Canberra ACT, 2601 Australia A thesis submitted in fulfilment of the requirements of the degree of Masters of Applied Science (Resource Management). August 2005. ii Abstract An ecological study was undertaken on four populations of Krefft’s River Turtle Emydura krefftii inhabiting the Townsville Area of Tropical North Queensland. Two sites were located in the Ross River, which runs through the urban areas of Townsville, and two sites were in rural areas at Alligator Creek and Stuart Creek (known as the Townsville Creeks). Earlier studies of the populations in Ross River had determined that the turtles existed at an exceptionally high density, that is, they were superabundant, and so the Townsville Creek sites were chosen as low abundance sites for comparison. The first aim of this study was to determine if there had been any demographic consequences caused by the abundance of turtle populations of the Ross River. Secondly, the project aimed to determine if the impoundments in the Ross River had affected the freshwater turtle fauna. Specifically this study aimed to determine if there were any difference between the growth, size at maturity, sexual dimorphism, size distribution, and diet of Emydura krefftii inhabiting two very different populations. A mark-recapture program estimated the turtle population sizes at between 490 and 5350 turtles per hectare. Most populations exhibited a predominant female sex-bias over the sampling period. Growth rates were rapid in juveniles but slowed once sexual maturity was attained; in males, growth basically stopped at maturity, but in females, growth continued post-maturity, although at a slower rate. -

Cultural Heritage Series
VOLUME 4 PART 2 MEMOIRS OF THE QUEENSLAND MUSEUM CULTURAL HERITAGE SERIES 17 OCTOBER 2008 © The State of Queensland (Queensland Museum) 2008 PO Box 3300, South Brisbane 4101, Australia Phone 06 7 3840 7555 Fax 06 7 3846 1226 Email [email protected] Website www.qm.qld.gov.au National Library of Australia card number ISSN 1440-4788 NOTE Papers published in this volume and in all previous volumes of the Memoirs of the Queensland Museum may be reproduced for scientific research, individual study or other educational purposes. Properly acknowledged quotations may be made but queries regarding the republication of any papers should be addressed to the Editor in Chief. Copies of the journal can be purchased from the Queensland Museum Shop. A Guide to Authors is displayed at the Queensland Museum web site A Queensland Government Project Typeset at the Queensland Museum CHAPTER 4 HISTORICAL MUA ANNA SHNUKAL Shnukal, A. 2008 10 17: Historical Mua. Memoirs of the Queensland Museum, Cultural Heritage Series 4(2): 61-205. Brisbane. ISSN 1440-4788. As a consequence of their different origins, populations, legal status, administrations and rates of growth, the post-contact western and eastern Muan communities followed different historical trajectories. This chapter traces the history of Mua, linking events with the family connections which always existed but were down-played until the second half of the 20th century. There are four sections, each relating to a different period of Mua’s history. Each is historically contextualised and contains discussions on economy, administration, infrastructure, health, religion, education and population. Totalai, Dabu, Poid, Kubin, St Paul’s community, Port Lihou, church missions, Pacific Islanders, education, health, Torres Strait history, Mua (Banks Island). -

Wetlands of the Townsville Area
A Final Report to the Townsville City Council WETLANDS OF THE TOWNSVILLE AREA ACTFR Report 96/28 25 November 1996 Prepared by G. Lukacs of the Australian Centre for Tropical Freshwater Research, James Cook University of North Queensland, Townsville Q 4811 Telephone (077 814262 Facsimile (077) 815589 Wetlands of the TCC LGA: Report No.96/28 TABLE OF CONTENTS 1. INTRODUCTION .................................................................................................................................. 1 1.1 Wetlands and the Community............................................................................................................. 1 1.2 The Wetlands of the Townsville Region ............................................................................................. 1 1.3 Values and Functions of Wetlands..................................................................................................... 3 2. METHODOLOGY ................................................................................................................................. 4 2.1 Scope .................................................................................................................................................. 4 2.2 Mapping ............................................................................................................................................. 4 2.3 Classification ..................................................................................................................................... 5 2.4 Sampling............................................................................................................................................ -

A Short History of Thuringowa
its 0#4, Wdkri Xdor# of fhurrngoraa Published by Thuringowa City Council P.O. Box 86, Thuringowa Central Queensland, 4817 Published October, 2000 Copyright The City of Thuringowa This book is copyright. Apart from any fair dealing for the purposes of private study, research, criticism or review, as permitted under the Copyright Act no part may be reproduced by any process without written permission. Inquiries should be addressed to the Publishers. All rights reserved. ISBN: 0 9577 305 3 5 kk THE CITY of Centenary of Federation i HURINGOWA Queensland This publication is a project initiated and funded by the City of Thuringowa This project is financially assisted by the Queensland Government, through the Queensland Community Assistance Program of the Centenary of Federation Queensland Cover photograph: Ted Gleeson crossing the Bohle. Gleeson Collection, Thuringowa Conienis Forward 5 Setting the Scene 7 Making the Land 8 The First People 10 People from the Sea 12 James Morrill 15 Farmers 17 Taking the Land 20 A Port for Thuringowa 21 Travellers 23 Miners 25 The Great Northern Railway 28 Growth of a Community 30 Closer Settlement 32 Towns 34 Sugar 36 New Industries 39 Empires 43 We can be our country 45 Federation 46 War in Europe 48 Depression 51 War in the North 55 The Americans Arrive 57 Prosperous Times 63 A great city 65 Bibliography 69 Index 74 Photograph Index 78 gOrtvard To celebrate our nations Centenary, and the various Thuringowan communities' contribution to our sense of nation, this book was commissioned. Two previous council publications, Thuringowa Past and Present and It Was a Different Town have been modest, yet tantalising introductions to facets of our past. -

Torres Strait Islanders: a New Deal
The Parliament of the Commonwealth of Australia TORRES STRAIT ISLANDERS: A NEW DEAL A REPORT ON GREATER AUTONOMY FOR TORRES STRAIT ISLANDERS House of Representatives Standing Committee on Aboriginal & Torres Strait Islander Affairs August 1997 Canberra Commonwealth of Australia 1997 ISBN This document was produced from camera-ready copy prepared by the House of Representatives Standing Committee on Aboriginal and Torres Strait Islander Affairs and printed by AGPS Canberra. The cover was produced in the AGPS design studios. The graphic on the cover was developed from a photograph taken on Yorke/Masig Island during the Committee's visit in October 1996. CONTENTS FOREWORD ix TERMS OF REFERENCE xii MEMBERSHIP OF THE COMMITTEE xiii GLOSSARY xiv SUMMARY AND RECOMMENDATIONS xv CHAPTER 1 – INTRODUCTION REFERRAL TO COMMITTEE.......................................................................................................................................1 CONDUCT OF THE INQUIRY ......................................................................................................................................1 SCOPE OF THE REPORT.............................................................................................................................................2 PRELIMINARY OBSERVATIONS .................................................................................................................................3 Commonwealth-State Cooperation ....................................................................................................................3 -
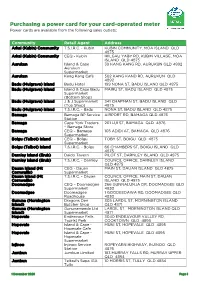
Card Operated Meter Information
Purchasing a power card for your card-operated meter Power cards are available from the following sales outlets: Community Retail Agent Address Arkai (Kubin) Community T.S.I.R.C. - Kubin KUBIN COMMUNITY, MOA ISLAND QLD 4875 Arkai (Kubin) Community CEQ - Kubin IKILGAU YABY RD, KUBIN VILLAGE, MOA ISLAND QLD 4875 Aurukun Island & Cape 39 KANG KANG RD, AURUKUN QLD 4892 Aurukun Supermarket Aurukun Kang Kang Café 502 KANG KAND RD, AURUKUN QLD 4892 Badu (Mulgrave) Island Badu Hotel 199 NONA ST, BADU ISLAND QLD 4875 Badu (Mulgrave) Island Island & Cape Badu MAIRU ST, BADU ISLAND QLD 4875 Supermarket (Bottom Shop) Badu (Mulgrave) Island J & J Supermarket 341 CHAPMAN ST, BADU ISLAND QLD (Top Shop) 4875 Badu (Mulgrave) Island T.S.I.R.C. - Badu NONA ST, BADU ISLAND QLD 4875 Bamaga Bamaga BP Service AIRPORT RD, BAMAGA QLD 4876 Station Bamaga Cape York Traders 201 LUI ST, BAMAGA QLD 4876 – Bamaga Store Bamaga CEQ – Bamaga 105 ADIDI AT, BAMAGA QLD 4876 Supermarket Boigu (Talbot) Island CEQ – Boigu TOBY ST, BOIGU QLD 4875 Supermarket Boigu (Talbot) Island T.S.I.R.C. - Boigu 66 CHAMBERS ST, BOIGU ISLAND QLD 4875 Darnley Island (Erub) Daido Tavern PILOT ST, DARNLEY ISLAND QLD 4875 Darnley Island (Erub) T.S.I.R.C. - Darnley COUNCIL OFFICE, DARNLEY ISLAND QLD 4875 Dauan Island (Mt CEQ - Dauan MAIN ST, DAUAN ISLAND QLD 4875 Cornwallis) Supermarket Dauan Island (Mt T.S.I.R.C. - Dauan COUNCIL OFFICE, MAIN ST, DAUAN Cornwallis) ISLAND QLD 4875 Doomadgee CEQ – Doomadgee 266 GUNNALUNJA DR, DOOMADGEE QLD Supermarket 4830 Doomadgee Doomadgee 1 GOODEEDAWA RD, DOOMADGEE -

WQ1181 - Ros! S River Basin
! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! Bohle River ! ! Stony Creek ! ! estuarine waters Town Common ! estuarine waters ! ! fresh waters ! ! ! ! ! Town Common ! ! ! estuarine waters ! ! ek ! e Cr ! ! ! r ! ! te a ! w ! ! ! *# e u ! l ek ! B re ! C ! ! ! ! ! ! ! ! y l a ! k e ! ! e r e Pallarenda fresh waters C H k ! ! ! ! ! e e ! ! r ! ! ! Deep C Cleveland Red Rock ! ! ! k ic l Pallarenda ! ! ! Bay A estuarine waters Ross Creek Bay ! ! Cape Cleveland ! estuarine waters ! ! fresh waters ! r ! ! e ! v ! ! i ! r R e ! v ! ! i R Rowes ! ! k c a e l l ! ! B Bay -

Badu Land and Sea Profile
Badu Land and Sea Profile RANGER GROUP Rangers 2015 MANAGEMENT PRIORITIES LAND • Native plants and animals • Burning• Land patrol • Weeds • Native nursery • Revegetation • Coastal management (beach patrol) OVERVIEW • Uninhabited island management Traditional island name Badu SEA • Crocodiles Western name Mulgrave • Turtle and dugong • Marine debris Western Islands Cluster Maluilgal Nation • Sea patrol • Seagrass Monitoring Local government TSIRC & TSC Registered Native Title Mura Badulgal (TSI) PEOPLE • Traditional ecological knowledge Body Corporate (RNTBC) Corporation RNTBC • Traditional and cultural sites • Community involvement Land type Continental Island • Visitor management Air distance from • Research support 49 Thursday Island (km) Area (ha) 10222 KEY VALUES Indicative max length (km) 11 CLIMATE CHANGE RISK Indicative max breadth (km) 13 Vulnerability to sea level rise (+1.0m) Very Low Max elevation (m) 190 Sea level rise response options Very High Healthy sea Marine water Coral reefs Seagrass Dugong Marine turtles Coastline length (km) 49 ecosystems quality meadows Population 783 (2011 ABS Census) Area of island zoned 162 development (ha) Subsistence Healthy land Sustainable Coasts Mangroves Coastal birds fishing ecosystems human settlements and beaches and wetlands Area of disturbed / 213 (2.1%) / undisturbed vegetation (ha/%) 10009 (97.9%) Supporting the Land and Sea Management Strategy for Torres Strait COMMUNITY OVERVIEW FUTURE SUSTAINABILITY INITIATIVES RECENT ACHIEVEMENTS Badu is a large (10,222ha) continental island in the The Badu community is highly reliant on air transport, Recent land and sea management achievements include: Near Western Cluster of the Torres Strait about diesel powered electricity generation and barge transport of supplies and materials to and from the community. 49km north of Thursday Island (Waiben). -
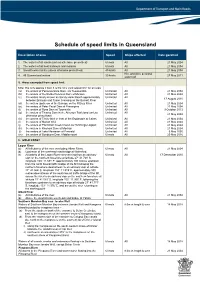
Schedule of Speed Limits in Queensland
Schedule of speed limits in Queensland Description of area Speed Ships affected Date gazetted 1. The waters of all canals (unless otherwise prescribed) 6 knots All 21 May 2004 2. The waters of all boat harbours and marinas 6 knots All 21 May 2004 3. Smooth water limits (unless otherwise prescribed) 40 knots All 21 May 2004 Hire and drive personal 4. All Queensland waters 30 knots 27 May 2011 watercraft 5. Areas exempted from speed limit Note: this only applies if item 3 is the only valid speed limit for an area (a) the waters of Perserverance Dam, via Toowoomba Unlimited All 21 May 2004 (b) the waters of the Bjelke Peterson Dam at Murgon Unlimited All 21 May 2004 (c) the waters locally known as Sandy Hook Reach approximately Unlimited All 17 August 2010 between Branyan and Tyson Crossing on the Burnett River (d) the waters upstream of the Barrage on the Fitzroy River Unlimited All 21 May 2004 (e) the waters of Peter Faust Dam at Proserpine Unlimited All 21 May 2004 (f) the waters of Ross Dam at Townsville Unlimited All 9 October 2013 (g) the waters of Tinaroo Dam in the Atherton Tableland (unless Unlimited All 21 May 2004 otherwise prescribed) (h) the waters of Trinity Inlet in front of the Esplanade at Cairns Unlimited All 21 May 2004 (i) the waters of Marian Weir Unlimited All 21 May 2004 (j) the waters of Plantation Creek known as Hutchings Lagoon Unlimited All 21 May 2004 (k) the waters in Kinchant Dam at Mackay Unlimited All 21 May 2004 (l) the waters of Lake Maraboon at Emerald Unlimited All 6 May 2005 (m) the waters of Bundoora Dam, Middlemount 6 knots All 20 May 2016 6.