Linear Algebra Has Some Conventional Ways of Representing Certain Types of Numerical Objects
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
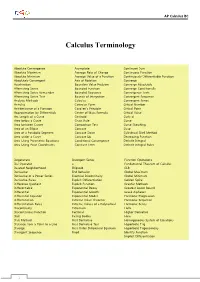
Calculus Terminology
AP Calculus BC Calculus Terminology Absolute Convergence Asymptote Continued Sum Absolute Maximum Average Rate of Change Continuous Function Absolute Minimum Average Value of a Function Continuously Differentiable Function Absolutely Convergent Axis of Rotation Converge Acceleration Boundary Value Problem Converge Absolutely Alternating Series Bounded Function Converge Conditionally Alternating Series Remainder Bounded Sequence Convergence Tests Alternating Series Test Bounds of Integration Convergent Sequence Analytic Methods Calculus Convergent Series Annulus Cartesian Form Critical Number Antiderivative of a Function Cavalieri’s Principle Critical Point Approximation by Differentials Center of Mass Formula Critical Value Arc Length of a Curve Centroid Curly d Area below a Curve Chain Rule Curve Area between Curves Comparison Test Curve Sketching Area of an Ellipse Concave Cusp Area of a Parabolic Segment Concave Down Cylindrical Shell Method Area under a Curve Concave Up Decreasing Function Area Using Parametric Equations Conditional Convergence Definite Integral Area Using Polar Coordinates Constant Term Definite Integral Rules Degenerate Divergent Series Function Operations Del Operator e Fundamental Theorem of Calculus Deleted Neighborhood Ellipsoid GLB Derivative End Behavior Global Maximum Derivative of a Power Series Essential Discontinuity Global Minimum Derivative Rules Explicit Differentiation Golden Spiral Difference Quotient Explicit Function Graphic Methods Differentiable Exponential Decay Greatest Lower Bound Differential -

History of Algebra and Its Implications for Teaching
Maggio: History of Algebra and its Implications for Teaching History of Algebra and its Implications for Teaching Jaime Maggio Fall 2020 MA 398 Senior Seminar Mentor: Dr.Loth Published by DigitalCommons@SHU, 2021 1 Academic Festival, Event 31 [2021] Abstract Algebra can be described as a branch of mathematics concerned with finding the values of unknown quantities (letters and other general sym- bols) defined by the equations that they satisfy. Algebraic problems have survived in mathematical writings of the Egyptians and Babylonians. The ancient Greeks also contributed to the development of algebraic concepts. In this paper, we will discuss historically famous mathematicians from all over the world along with their key mathematical contributions. Mathe- matical proofs of ancient and modern discoveries will be presented. We will then consider the impacts of incorporating history into the teaching of mathematics courses as an educational technique. 1 https://digitalcommons.sacredheart.edu/acadfest/2021/all/31 2 Maggio: History of Algebra and its Implications for Teaching 1 Introduction In order to understand the way algebra is the way it is today, it is important to understand how it came about starting with its ancient origins. In a mod- ern sense, algebra can be described as a branch of mathematics concerned with finding the values of unknown quantities defined by the equations that they sat- isfy. Algebraic problems have survived in mathematical writings of the Egyp- tians and Babylonians. The ancient Greeks also contributed to the development of algebraic concepts, but these concepts had a heavier focus on geometry [1]. The combination of all of the discoveries of these great mathematicians shaped the way algebra is taught today. -

The Evolution of Equation-Solving: Linear, Quadratic, and Cubic
California State University, San Bernardino CSUSB ScholarWorks Theses Digitization Project John M. Pfau Library 2006 The evolution of equation-solving: Linear, quadratic, and cubic Annabelle Louise Porter Follow this and additional works at: https://scholarworks.lib.csusb.edu/etd-project Part of the Mathematics Commons Recommended Citation Porter, Annabelle Louise, "The evolution of equation-solving: Linear, quadratic, and cubic" (2006). Theses Digitization Project. 3069. https://scholarworks.lib.csusb.edu/etd-project/3069 This Thesis is brought to you for free and open access by the John M. Pfau Library at CSUSB ScholarWorks. It has been accepted for inclusion in Theses Digitization Project by an authorized administrator of CSUSB ScholarWorks. For more information, please contact [email protected]. THE EVOLUTION OF EQUATION-SOLVING LINEAR, QUADRATIC, AND CUBIC A Project Presented to the Faculty of California State University, San Bernardino In Partial Fulfillment of the Requirements for the Degre Master of Arts in Teaching: Mathematics by Annabelle Louise Porter June 2006 THE EVOLUTION OF EQUATION-SOLVING: LINEAR, QUADRATIC, AND CUBIC A Project Presented to the Faculty of California State University, San Bernardino by Annabelle Louise Porter June 2006 Approved by: Shawnee McMurran, Committee Chair Date Laura Wallace, Committee Member , (Committee Member Peter Williams, Chair Davida Fischman Department of Mathematics MAT Coordinator Department of Mathematics ABSTRACT Algebra and algebraic thinking have been cornerstones of problem solving in many different cultures over time. Since ancient times, algebra has been used and developed in cultures around the world, and has undergone quite a bit of transformation. This paper is intended as a professional developmental tool to help secondary algebra teachers understand the concepts underlying the algorithms we use, how these algorithms developed, and why they work. -

Mathematics for Earth Science
Mathematics for Earth Science The module covers concepts such as: • Maths refresher • Fractions, Percentage and Ratios • Unit conversions • Calculating large and small numbers • Logarithms • Trigonometry • Linear relationships Mathematics for Earth Science Contents 1. Terms and Operations 2. Fractions 3. Converting decimals and fractions 4. Percentages 5. Ratios 6. Algebra Refresh 7. Power Operations 8. Scientific Notation 9. Units and Conversions 10. Logarithms 11. Trigonometry 12. Graphs and linear relationships 13. Answers 1. Terms and Operations Glossary , 2 , 3 & 17 are TERMS x4 + 2 + 3 = 17 is an EQUATION 17 is the SUM of + 2 + 3 4 4 4 is an EXPONENT + 2 + 3 = 17 4 3 is a CONSTANT 2 is a COEFFICIENT is a VARIABLE + is an OPERATOR +2 + 3 is an EXPRESSION 4 Equation: A mathematical sentence containing an equal sign. The equal sign demands that the expressions on either side are balanced and equal. Expression: An algebraic expression involves numbers, operation signs, brackets/parenthesis and variables that substitute numbers but does not include an equal sign. Operator: The operation (+ , ,× ,÷) which separates the terms. Term: Parts of an expression− separated by operators which could be a number, variable or product of numbers and variables. Eg. 2 , 3 & 17 Variable: A letter which represents an unknown number. Most common is , but can be any symbol. Constant: Terms that contain only numbers that always have the same value. Coefficient: A number that is partnered with a variable. The term 2 is a coefficient with variable. Between the coefficient and variable is a multiplication. Coefficients of 1 are not shown. Exponent: A value or base that is multiplied by itself a certain number of times. -

Algebra Vocabulary List (Definitions for Middle School Teachers)
Algebra Vocabulary List (Definitions for Middle School Teachers) A Absolute Value Function – The absolute value of a real number x, x is ⎧ xifx≥ 0 x = ⎨ ⎩−<xifx 0 http://www.math.tamu.edu/~stecher/171/F02/absoluteValueFunction.pdf Algebra Lab Gear – a set of manipulatives that are designed to represent polynomial expressions. The set includes representations for positive/negative 1, 5, 25, x, 5x, y, 5y, xy, x2, y2, x3, y3, x2y, xy2. The manipulatives can be used to model addition, subtraction, multiplication, division, and factoring of polynomials. They can also be used to model how to solve linear equations. o For more info: http://www.stlcc.cc.mo.us/mcdocs/dept/math/homl/manip.htm http://www.awl.ca/school/math/mr/alg/ss/series/algsrtxt.html http://www.picciotto.org/math-ed/manipulatives/lab-gear.html Algebra Tiles – a set of manipulatives that are designed for modeling algebraic expressions visually. Each tile is a geometric model of a term. The set includes representations for positive/negative 1, x, and x2. The manipulatives can be used to model addition, subtraction, multiplication, division, and factoring of polynomials. They can also be used to model how to solve linear equations. o For more info: http://math.buffalostate.edu/~it/Materials/MatLinks/tilelinks.html http://plato.acadiau.ca/courses/educ/reid/Virtual-manipulatives/tiles/tiles.html Algebraic Expression – a number, variable, or combination of the two connected by some mathematical operation like addition, subtraction, multiplication, division, exponents and/or roots. o For more info: http://www.wtamu.edu/academic/anns/mps/math/mathlab/beg_algebra/beg_alg_tut11 _simp.htm http://www.math.com/school/subject2/lessons/S2U1L1DP.html Area Under the Curve – suppose the curve y=f(x) lies above the x axis for all x in [a, b]. -

Polynomial Coefficient Enumeration
Polynomial Coefficient Enumeration Tewodros Amdeberhan∗ Richard P. Stanley† February 03, 2008 Abstract Let f(x1, . , xk) be a polynomial over a field K. This paper con- siders such questions as the enumeration of the number of nonzero coefficients of ∗ f or of the number of coefficients equal to α ∈ K . For instance, if K = Fq then a matrix formula is obtained for the number of coefficients of f n that are equal to ∗ α ∈ Fq, as a function of n. Many additional results are obtained related to such areas as lattice path enumeration and the enumeration of integer points in convex polytopes. 1 Introduction. Given a polynomial f ∈ Z[x1, . , xn], how many coefficients of f are nonzero? For α ∈ Z and p prime, how many coefficients are congruent to α modulo p? In this paper we will investigate these and related questions. First let us review some known results that will suggest various generalizations. The archetypal result for understanding the coefficients of a polynomial modulo p is Lucas’ theorem [7]: if n, k are positive integers with p-ary ex- P i P i pansions n = i≥0 aip and k = i≥0 bip , then n a a ≡ 0 1 ··· (mod p). k b0 b1 ∗Department of Mathematics, Massachusetts Institute of Technology, Cambridge, MA 02139 Email: [email protected] †Department of Mathematics, Massachusetts Institute of Technology, Cambridge, MA 02139 Email: [email protected] 1 Thus, for instance, it immediately follows that the number of odd coefficients of the polynomial (1 + x)n is equal to 2s(n), where s(n) denotes the number of 1’s in the 2-ary (binary) expansion of n. -

Massachusetts Mathematics Curriculum Framework — 2017
Massachusetts Curriculum MATHEMATICS Framework – 2017 Grades Pre-Kindergarten to 12 i This document was prepared by the Massachusetts Department of Elementary and Secondary Education Board of Elementary and Secondary Education Members Mr. Paul Sagan, Chair, Cambridge Mr. Michael Moriarty, Holyoke Mr. James Morton, Vice Chair, Boston Dr. Pendred Noyce, Boston Ms. Katherine Craven, Brookline Mr. James Peyser, Secretary of Education, Milton Dr. Edward Doherty, Hyde Park Ms. Mary Ann Stewart, Lexington Dr. Roland Fryer, Cambridge Mr. Nathan Moore, Chair, Student Advisory Council, Ms. Margaret McKenna, Boston Scituate Mitchell D. Chester, Ed.D., Commissioner and Secretary to the Board The Massachusetts Department of Elementary and Secondary Education, an affirmative action employer, is committed to ensuring that all of its programs and facilities are accessible to all members of the public. We do not discriminate on the basis of age, color, disability, national origin, race, religion, sex, or sexual orientation. Inquiries regarding the Department’s compliance with Title IX and other civil rights laws may be directed to the Human Resources Director, 75 Pleasant St., Malden, MA, 02148, 781-338-6105. © 2017 Massachusetts Department of Elementary and Secondary Education. Permission is hereby granted to copy any or all parts of this document for non-commercial educational purposes. Please credit the “Massachusetts Department of Elementary and Secondary Education.” Massachusetts Department of Elementary and Secondary Education 75 Pleasant Street, Malden, MA 02148-4906 Phone 781-338-3000 TTY: N.E.T. Relay 800-439-2370 www.doe.mass.edu Massachusetts Department of Elementary and Secondary Education 75 Pleasant Street, Malden, Massachusetts 02148-4906 Dear Colleagues, I am pleased to present to you the Massachusetts Curriculum Framework for Mathematics adopted by the Board of Elementary and Secondary Education in March 2017. -

Eigenvalues and Eigenvectors
Chapter 3 Eigenvalues and Eigenvectors In this chapter we begin our study of the most important, and certainly the most dominant aspect, of matrix theory. Called spectral theory, it allows us to give fundamental structure theorems for matrices and to develop power tools for comparing and computing withmatrices.Webeginwithastudy of norms on matrices. 3.1 Matrix Norms We know Mn is a vector space. It is most useful to apply a metric on this vector space. The reasons are manifold, ranging from general information of a metrized system to perturbation theory where the “smallness” of a matrix must be measured. For that reason we define metrics called matrix norms that are regular norms with one additional property pertaining to the matrix product. Definition 3.1.1. Let A Mn.Recallthatanorm, ,onanyvector space satifies the properties:∈ · (i) A 0and A =0ifandonlyifA =0 ≥ | (ii) cA = c A for c R | | ∈ (iii) A + B A + B . ≤ There is a true vector product on Mn defined by matrix multiplication. In this connection we say that the norm is submultiplicative if (iv) AB A B ≤ 95 96 CHAPTER 3. EIGENVALUES AND EIGENVECTORS In the case that the norm satifies all four properties (i) - (iv) we call it a matrix norm. · Here are a few simple consequences for matrix norms. The proofs are straightforward. Proposition 3.1.1. Let be a matrix norm on Mn, and suppose that · A Mn.Then ∈ (a) A 2 A 2, Ap A p,p=2, 3,... ≤ ≤ (b) If A2 = A then A 1 ≥ 1 I (c) If A is invertible, then A− A ≥ (d) I 1. -

A Universal Coefficient Theorem with Applications to Torsion in Chow Groups of Severi–Brauer Varieties
New York Journal of Mathematics New York J. Math. 26 (2020) 1155{1183. A universal coefficient theorem with applications to torsion in Chow groups of Severi{Brauer varieties Eoin Mackall Abstract. For any variety X, and for any coefficient ring S, we de- fine the S-topological filtration on the Grothendieck group of coherent sheaves G(X) ⊗ S with coefficients in S. The S-topological filtration is related to the topological filtration by means of a universal coefficient theorem. We apply this observation in the case X is a Severi{Brauer variety to obtain new examples of torsion in the Chow groups of X. Contents 1. Introduction 1155 2. Filtered rings with coefficients 1157 3. The topological filtration with coefficients 1159 4. Generic algebras of index 2; 4; 8; 16 and 32 1164 References 1182 Notation and Conventions. A ring is a commutative ring. An abelian group is flat if it is a flat Z-module; equivalently, an abelian group is flat if and only if it is torsion free. We fix an arbitrary field k, to be used as a base. For any field F , an F -variety (or simply a variety when the field F is clear) is an integral scheme separated and of finite type over F . 1. Introduction Let X be a Severi{Brauer variety. The Chow groups CHi(X) of algebraic cycles on X of codimension-i modulo rational equivalence have been the subject of a considerable amount of current research [1,9, 10, 11, 12]. A primary focus of this research has been to determine the possible torsion subgroups of CHi(X) for varying i and X. -

Vector Spaces
Chapter 2 Vector Spaces 2.1 INTRODUCTION In various practical and theoretical problems, we come across a set V whose elements may be vectors in two or three dimensions or real-valued functions, which can be added and multi- plied by a constant (number) in a natural way, the result being again an element of V. Such concrete situations suggest the concept of a vector space. Vector spaces play an important role in many branches of mathematics and physics. In analysis infinite dimensional vector spaces (in fact, normed vector spaces) are more important than finite dimensional vector spaces while in linear algebra finite dimensional vector spaces are used, because they are simple and linear transformations on them can be represented by matrices. This chapter mainly deals with finite dimensional vector spaces. 2.2 DEFINITION AND EXAMPLES VECTOR SPACE. An algebraic structure (V, F, ⊕, ) consisting of a non-void set V , a field F, a binary operation ⊕ on V and an external mapping : F ×V → V associating each a ∈ F,v ∈ V to a unique element a v ∈ V is said to be a vector space over field F, if the following axioms are satisfied: V -1 (V, ⊕) is an abelian group. V -2 For all u, v ∈ V and a, b ∈ F, we have (i) a (u ⊕ v)=a u ⊕ a v, (ii) (a + b) u = a u ⊕ b u, (iii) (ab) u = a (b u), (iv) 1 u = u. The elements of V are called vectors and those of F are called scalars. The mapping is called scalar multiplication and the binary operation ⊕ is termed vector addition. -

SOME PROPERTIES of the COEFFICIENT of VARIATION and F STATISTICS with RESPECT to TRANSFORMATIONS of the FORM Xk*
SOME PROPERTIES OF THE COEFFICIENT OF VARIATION AND F STATISTICS WITH RESPECT TO TRANSFORMATIONS OF THE FORM Xk* B. M. Rao a...nd W. T. Federer I. INTRODUCTION The consequences of non-normality, non-additivity, and heterogeneity of error variances on F tests in the analysis of variance have been studied in a n~~ber of papers (e.g., Bartlett [1,2], Box and Anderson [3], Cochran [4,5], Cu.:ctiss [6], Harter and lJ..l!a [7], Nissen and Ottestad [8), Pearson [9], Tuk.ey [10]). It appears that some of the conclusions drawn need fill~ther investiga.- tion. Also, a statistical ~rocedure designed "t9 determine the scale of measure- ment required to obtain any of these conditions is not available. Tukey [11] has n~e some progress in this direction; ~he statistic designed to determine the appropriate scale of measurement for a set of data to obtain normality is u.navaila.ble • In an attempt to firrl such a statistic, and in vievT of the folloWing comment made by Pearson [9] "• • • But in the more extrem~e cases of non- no~mal variation, where the standard deviation becomes an unsatisfactory measure of variability, there will always be a danger of accepting the hypo· to pick out a real difference,", it was decided to investigate the minimum coefficient of variation (C.V), a minimum F ratio for Tukey 1s one degree of freedom for non-additivity, and a maximum F ratios for row and column mean squares. Some ~nalytic results were obtained for the coefficient of variation in relation to positive integer values of k in the transforma- tion xt. -

Glossary and Notation
A Glossary and Notation AB: Adams–Bashforth—names of a family of explicit LMMs. AB(2) denotes the two-step, second-order Adams–Bashforth method. AM: Adams–Moulton—names of a family of implicit LMMs. AM(2) denotes the two-step, third-order Adams–Moulton method. BDF: backward differentiation formula. BE: backward Euler (method). Cp+1 : error constant of a pth-order LMM. CF: complementary function—the general solution of a homogeneous linear difference or differential equation. 4E: difference equation. E(·): expected value. fn : the value of f(t, x) at t = tn and x = xn. FE: forward Euler (method). GE: global error—the difference between the exact solution x(tn) at t = tn and the numerical solution xn: en = x(tn) − xn. ∇: gradient operator. ∇F denotes the vector of partial derivatives of F. h: step size—numerical solutions are sought at times tn = t0 + nh for n = 0, 1, 2,... D.F. Griffiths, D.J. Higham, Numerical Methods for Ordinary Differential Equations, Springer Undergraduate Mathematics Series, DOI 10.1007/978-0-85729-148-6, © Springer-Verlag London Limited 2010 244 A. Glossary and Notation bh : λh, where λ is the coefficient in the first-order equation x0(t) = λx(t) or an eigenvalue of the matrix A in the system of ODEs x0(t) = Ax(t). I: identity matrix. =(λ): imaginary part of a complex number λ. IAS: interval of absolute stability. IC: initial condition. IVP: initial value problem—an ODE together with initial condition(s). j = m : n: for integers m < n this is shorthand for the sequence of consecutive integers from m to n.