Functional Segregation of the Human Basal Forebrain Using Resting State Neuroimaging
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synaptic Organization of Claustral and Geniculate Afferents to the Visual Cortex of the Cat
The Journal of Neuroscience December 1986, 6(12): 3564-3575 Synaptic Organization of Claustral and Geniculate Afferents to the Visual Cortex of the Cat Simon LeVay Robert Bosch Vision Research Center, Salk Institute for Biological Studies, San Diego, California 92138 Claustral and geniculate afferents to area 17 were labeled by sponses of some cortical neurons, previously called “hypercom- anterograde axonal transport of peroxidase-conjugated wheat- plex cells” by Hubel and Wiesel (1965), are suppressed as the germ agglutinin, and examined in the electron microscope. A length of the stimulating slit of light is extended beyond some peroxidase reaction protocol that led to labeling in the form of optimal value. Interestingly, neurons in the visual claustrum minute holes in the EM sections was used. Both types of affer- behave in a complementary fashion: Their responses to an ori- ents formed type 1 (presumed excitatory) synapses exclusively. ented stimulus increase monotonically with stimulus length, In agreement with previous reports the great majority of genic- sometimes showing length summation up to 40” or more-a ulate afferents to layers 4 and 6 contacted dendritic spines. The sizable portion of the animal’s entire field of view (Sherk and claustral afferents to layers 1 and 6 also predominantly con- LeVay, 198 1). tacted spines. In layer 4, however, claustral afferents contacted These observations suggest that claustral neurons contribute spines and dendritic shafts about equally. The results suggest a to end-stopping by exerting a length-dependent inhibition on substantial Claus&al input to smooth-dendrite cells in layer 4, some neurons in the visual cortex. -

The Effect of Nucleus Basalis Magnocellularis Deep Brain
Lee et al. BMC Neurology (2016) 16:6 DOI 10.1186/s12883-016-0529-z RESEARCH ARTICLE Open Access The effect of nucleus basalis magnocellularis deep brain stimulation on memory function in a rat model of dementia Ji Eun Lee1†, Da Un Jeong1†, Jihyeon Lee1, Won Seok Chang2 and Jin Woo Chang1,2* Abstract Background: Deep brain stimulation has recently been considered a potential therapy in improving memory function. It has been shown that a change of neurotransmitters has an effect on memory function. However, much about the exact underlying neural mechanism is not yet completely understood. We therefore examined changes in neurotransmitter systems and spatial memory caused by stimulation of nucleus basalis magnocellularis in a rat model of dementia. Methods: We divided rats into four groups: Normal, Lesion, Implantation, and Stimulation. We used 192 IgG-saporin for degeneration of basal forebrain cholinergic neuron related with learning and memory and it was injected into all rats except for the normal group. An electrode was ipsilaterally inserted in the nucleus basalis magnocellularis of all rats of the implantation and stimulation group, and the stimulation group received the electrical stimulation. Features were verified by the Morris water maze, immunochemistry and western blotting. Results: AllgroupsshowedsimilarperformancesduringMorriswater maze training. During the probe trial, performance of the lesion and implantation group decreased. However, the stimulation group showed an equivalent performance to the normal group. In the lesion and implantation group, expression of glutamate acid decarboxylase65&67 decreased in the medial prefrontal cortex and expression of glutamate transporters increased in the medial prefrontal cortex and hippocampus. However, expression of the stimulation group showed similar levels as the normal group. -

Neural Activity in the Horizontal Limb of the Diagonal Band of Broca Can Be Modulated by Electrical Stimulation of the Olfactory Bulb and Cortex in Rats
Neuroscience Letters 282 (2000) 157±160 www.elsevier.com/locate/neulet Neural activity in the horizontal limb of the diagonal band of Broca can be modulated by electrical stimulation of the olfactory bulb and cortex in rats Christiane Linster*, Michael E. Hasselmo Department of Psychology, Boston University, 64 Cummington Street, Boston, MA 02215, USA Received 10 January 2000; received in revised form 31 January 2000; accepted 1 February 2000 Abstract Previously published theoretical models of olfactory processing suggest that cholinergic modulatory inputs to the olfactory system should be regulated by neural activity in the olfactory bulb. We tested these predictions using in vivo electrophysiology in rats. We show that the activity of approximately 20% of neurons recorded in the horizontal limb of the diagonal band of Broca (HDB), which is the source of cholinergic projections to the olfactory system, can be modulated by electrical stimulation of either the lateral olfactory tract or the cell body layer of piriform cortex. These data suggest a possible physiological pathway for the proposed regulation of neural activity in the HDB by activity in the olfactory bulb or cortex. q 2000 Elsevier Science Ireland Ltd. All rights reserved. Keywords: Olfactory bulb; Olfactory cortex; Horizontal limb of the diagonal band of Broca; Cholinergic modulation; Electrical stimula- tion; Units recordings Understanding the role of neuromodulators for cortical brain structure [23]. The HDB contains both cholinergic and function requires study of their effects at the level of neural GABAergic neurons, and it is known that these two popula- activity as well as at the level of behavioral responses. -

Fos Activation of Selective Afferents to Ventral Tegmental Area During Cue-Induced Reinstatement of Cocaine Seeking in Rats
The Journal of Neuroscience, September 19, 2012 • 32(38):13309–13325 • 13309 Behavioral/Systems/Cognitive Fos Activation of Selective Afferents to Ventral Tegmental Area during Cue-Induced Reinstatement of Cocaine Seeking in Rats Stephen V. Mahler and Gary S. Aston-Jones Department of Neurosciences, Medical University of South Carolina, Charleston, South Carolina 29425 Ventral tegmental area (VTA) dopamine neurons are crucial for appetitive responses to Pavlovian cues, including cue-induced reinstate- ment of drug seeking. However, it is unknown which VTA inputs help activate these neurons, transducing stimuli into salient cues that drive drug-seeking behavior. Here we examined 56 VTA afferents from forebrain and midbrain that are Fos activated during cue-induced reinstatement. We injected the retrograde tracer cholera toxin  subunit (CTb) unilaterally into rostral or caudal VTA of male rats. All animalsweretrainedtoself-administercocaine,thenextinguishedofthisbehavior.Onafinaltestday,animalswereexposedtoresponse- contingent cocaine-associated cues, extinction conditions, a non-cocaine-predictive CSϪ, or a novel environment, and brains were processed to visualize CTb and Fos immunoreactivity to identify VTA afferents activated in relation to behaviors. VTA-projecting neurons in subregions of medial accumbens shell, ventral pallidum, elements of extended amygdala, and lateral septum (but not pre- frontal cortex) were activated specifically during cue-induced cocaine seeking, and some of these were also activated proportionately to the degree of cocaine seeking. Surprisingly, though efferents from the lateral hypothalamic orexin field were also Fos activated during reinstatement, these were largely non-orexinergic. Also, VTA afferents from the rostromedial tegmental nucleus and lateral habenula were specifically activated during extinction and CSϪ tests, when cocaine was not expected. -

Implications for Learning and Memory
The Journal of Neuroscience, May 15, 2000, 20(10):3900–3908 Cholinergic Excitation of Septohippocampal GABA But Not Cholinergic Neurons: Implications for Learning and Memory Min Wu,1 Marya Shanabrough,2 Csaba Leranth,2,3 and Meenakshi Alreja1,3 Departments of 1Psychiatry, 2Obstetrics and Gynecology, and 3Neurobiology, Yale University School of Medicine and the Ribicoff Research Facilities, Connecticut Mental Health Center, New Haven, Connecticut 06508 The medial septum/diagonal band (MSDB), which gives rise to linergic neurons in rat brain slices, we have found that musca- the septohippocampal pathway, is a critical locus for the mne- rinic agonists do not excite septohippocampal cholinergic neu- monic effects of muscarinic drugs. Infusion of muscarinic cho- rons, instead they inhibit a subpopulation of cholinergic linergic agonists into the MSDB enhance learning and memory neurons. In contrast, unlabeled neurons, confirmed to be non- processes both in young and aged rats and produce a contin- cholinergic, septohippocampal GABA-type neurons using ret- uous theta rhythm in the hippocampus. Intraseptal muscarinic rograde marking and double-labeling techniques, are pro- agonists also alleviate the amnesic syndrome produced by foundly excited by muscarine. Thus, the cognition-enhancing systemic administration of muscarinic receptor antagonists. It effects of muscarinic drugs in the MSDB cannot be attributed to has been presumed, but not proven, that the cellular mecha- an increase in hippocampal ACh release. Instead, disinhibitory nisms underlying the effects of muscarinic agonists in the mechanisms, caused by increased impulse flow in the septo- MSDB involve an excitation of septohippocampal cholinergic hippocampal GABAergic pathway, may underlie the cognition- neurons and a subsequent increase in acetylcholine (ACh) re- enhancing effects of muscarinic agonists. -

Intersection of Structural and Functional Connectivity of the Nucleus Basalis of Meynert in Parkinson's Disease Dementia and L
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.27.221853; this version posted July 28, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Intersection of structural and functional connectivity of the nucleus basalis of Meynert in Parkinson’s disease dementia and Lewy body dementia. Authors: Ashwini Oswala,b,c*†, James Gratwicked†, Harith Akramd, Marjan Jahanshahid, Laszlo Zaborszkye, Peter Browna,b , Marwan Harizd,f, Ludvic Zrinzod, Tom Foltynied, Vladimir Litvakc †Denotes equal contribution to Authorship Affiliations: a Medical Research Brain Network Dynamics Unit, University of Oxford, Oxford, UK b Nuffield Department of Clinical Neurosciences, John Radcliffe Hospital, Oxford, UK c Wellcome Trust Centre for Neuroimaging, UCL Institute of Neurology, 12 Queen Square, London, UK d Department of Clinical & Movement Neurosciences, UCL Institute of Neurology and The National Hospital for Neurology and Neurosurgery, Queen Square, London, UK e Center for Molecular and Behavioral Neuroscience, Rutgers University, USA f Department of Clinical Neuroscience, Umeå University, Umeå, Sweden To whom correspondence should be addressed: [email protected] or [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.07.27.221853; this version posted July 28, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. -
Lecture 12 Notes
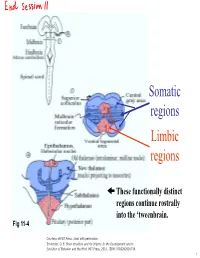
Somatic regions Limbic regions These functionally distinct regions continue rostrally into the ‘tweenbrain. Fig 11-4 Courtesy of MIT Press. Used with permission. Schneider, G. E. Brain structure and its Origins: In the Development and in Evolution of Behavior and the Mind. MIT Press, 2014. ISBN: 9780262026734. 1 Chapter 11, questions about the somatic regions: 4) There are motor neurons located in the midbrain. What movements do those motor neurons control? (These direct outputs of the midbrain are not a subject of much discussion in the chapter.) 5) At the base of the midbrain (ventral side) one finds a fiber bundle that shows great differences in relative size in different species. Give examples. What are the fibers called and where do they originate? 8) A decussating group of axons called the brachium conjunctivum also varies greatly in size in different species. It is largest in species with the largest neocortex but does not come from the neocortex. From which structure does it come? Where does it terminate? (Try to guess before you look it up.) 2 Motor neurons of the midbrain that control somatic muscles: the oculomotor nuclei of cranial nerves III and IV. At this level, the oculomotor nucleus of nerve III is present. Fibers from retina to Superior Colliculus Brachium of Inferior Colliculus (auditory pathway to thalamus, also to SC) Oculomotor nucleus Spinothalamic tract (somatosensory; some fibers terminate in SC) Medial lemniscus Cerebral peduncle: contains Red corticospinal + corticopontine fibers, + cortex to hindbrain fibers nucleus (n. ruber) Tectospinal tract Rubrospinal tract Courtesy of MIT Press. Used with permission. Schneider, G. -

Memory Loss from a Subcortical White Matter Infarct
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.51.6.866 on 1 June 1988. Downloaded from Journal of Neurology, Neurosurgery, and Psychiatry 1988;51:866-869 Short report Memory loss from a subcortical white matter infarct CAROL A KOOISTRA, KENNETH M HEILMAN From the Department ofNeurology, College ofMedicine, University ofFlorida, and the Research Service, Veterans Administration Medical Center, Gainesville, FL, USA SUMMARY Clinical disorders of memory are believed to occur from the dysfunction of either the mesial temporal lobe, the mesial thalamus, or the basal forebrain. Fibre tract damage at the level of the fornix has only inconsistently produced amnesia. A patient is reported who suffered a cerebro- vascular accident involving the posterior limb of the left internal capsule that resulted in a persistent and severe disorder of verbal memory. The inferior extent of the lesion effectively disconnected the mesial thalamus from the amygdala and the frontal cortex by disrupting the ventral amygdalofugal and thalamic-frontal pathways as they course through the diencephalon. This case demonstrates that an isolated lesion may cause memory loss without involvement of traditional structures associated with memory and may explain memory disturbances in other white matter disease such as multiple sclerosis and lacunar state. Protected by copyright. Memory loss is currently believed to reflect grey day of his illness the patient was transferred to Shands matter damage of either the mesial temporal lobe,' -4 Teaching Hospital at the University of Florida for further the mesial or the basal forebrain.'0 l evaluation. thalamus,5-9 Examination at that time showed the patient to be awake, Cerebrovascular accidents resulting in memory dys- alert, attentive and fully oriented. -

Pallial Origin of Basal Forebrain Cholinergic Neurons in the Nucleus
ERRATUM 4565 Development 138, 4565 (2011) doi:10.1242/dev.074088 © 2011. Published by The Company of Biologists Ltd Pallial origin of basal forebrain cholinergic neurons in the nucleus basalis of Meynert and horizontal limb of the diagonal band nucleus Ana Pombero, Carlos Bueno, Laura Saglietti, Monica Rodenas, Jordi Guimera, Alexandro Bulfone and Salvador Martinez There was an error in the ePress version of Development 138, 4315-4326 published on 24 August 2011. In Fig. 7P, the P-values are not given in the legend. For region 2, P=0.02; for region 3, P=0.003. The final online issue and print copy are correct. We apologise to authors and readers for this error. DEVELOPMENT RESEARCH ARTICLE 4315 Development 138, 4315-4326 (2011) doi:10.1242/dev.069534 © 2011. Published by The Company of Biologists Ltd Pallial origin of basal forebrain cholinergic neurons in the nucleus basalis of Meynert and horizontal limb of the diagonal band nucleus Ana Pombero1, Carlos Bueno1, Laura Saglietti2, Monica Rodenas1, Jordi Guimera3, Alexandro Bulfone4 and Salvador Martinez1,* SUMMARY The majority of the cortical cholinergic innervation implicated in attention and memory originates in the nucleus basalis of Meynert and in the horizontal limb of the diagonal band nucleus of the basal prosencephalon. Functional alterations in this system give rise to neuropsychiatric disorders as well as to the cognitive alterations described in Parkinson and Alzheimer’s diseases. Despite the functional importance of these basal forebrain cholinergic neurons very little is known about their origin and development. Previous studies suggest that they originate in the medial ganglionic eminence of the telencephalic subpallium; however, our results identified Tbr1-expressing, reelin-positive neurons migrating from the ventral pallium to the subpallium that differentiate into cholinergic neurons in the basal forebrain nuclei projecting to the cortex. -

The Three Amnesias
The Three Amnesias Russell M. Bauer, Ph.D. Department of Clinical and Health Psychology College of Public Health and Health Professions Evelyn F. and William L. McKnight Brain Institute University of Florida PO Box 100165 HSC Gainesville, FL 32610-0165 USA Bauer, R.M. (in press). The Three Amnesias. In J. Morgan and J.E. Ricker (Eds.), Textbook of Clinical Neuropsychology. Philadelphia: Taylor & Francis/Psychology Press. The Three Amnesias - 2 During the past five decades, our understanding of memory and its disorders has increased dramatically. In 1950, very little was known about the localization of brain lesions causing amnesia. Despite a few clues in earlier literature, it came as a complete surprise in the early 1950’s that bilateral medial temporal resection caused amnesia. The importance of the thalamus in memory was hardly suspected until the 1970’s and the basal forebrain was an area virtually unknown to clinicians before the 1980’s. An animal model of the amnesic syndrome was not developed until the 1970’s. The famous case of Henry M. (H.M.), published by Scoville and Milner (1957), marked the beginning of what has been called the “golden age of memory”. Since that time, experimental analyses of amnesic patients, coupled with meticulous clinical description, pathological analysis, and, more recently, structural and functional imaging, has led to a clearer understanding of the nature and characteristics of the human amnesic syndrome. The amnesic syndrome does not affect all kinds of memory, and, conversely, memory disordered patients without full-blown amnesia (e.g., patients with frontal lesions) may have impairment in those cognitive processes that normally support remembering. -

Distribution of Dopamine D3 Receptor Expressing Neurons in the Human Forebrain: Comparison with D2 Receptor Expressing Neurons Eugenia V
Distribution of Dopamine D3 Receptor Expressing Neurons in the Human Forebrain: Comparison with D2 Receptor Expressing Neurons Eugenia V. Gurevich, Ph.D., and Jeffrey N. Joyce, Ph.D. The dopamine D2 and D3 receptors are members of the D2 important difference from the rat is that D3 receptors were subfamily that includes the D2, D3 and D4 receptor. In the virtually absent in the ventral tegmental area. D3 receptor rat, the D3 receptor exhibits a distribution restricted to and D3 mRNA positive neurons were observed in sensory, mesolimbic regions with little overlap with the D2 receptor. hormonal, and association regions such as the nucleus Receptor binding and nonisotopic in situ hybridization basalis, anteroventral, mediodorsal, and geniculate nuclei of were used to study the distribution of the D3 receptors and the thalamus, mammillary nuclei, the basolateral, neurons positive for D3 mRNA in comparison to the D2 basomedial, and cortical nuclei of the amygdala. As revealed receptor/mRNA in subcortical regions of the human brain. by simultaneous labeling for D3 and D2 mRNA, D3 mRNA D2 binding sites were detected in all brain areas studied, was often expressed in D2 mRNA positive neurons. with the highest concentration found in the striatum Neurons that solely expressed D2 mRNA were numerous followed by the nucleus accumbens, external segment of the and regionally widespread, whereas only occasional D3- globus pallidus, substantia nigra and ventral tegmental positive-D2-negative cells were observed. The regions of area, medial preoptic area and tuberomammillary nucleus relatively higher expression of the D3 receptor and its of the hypothalamus. In most areas the presence of D2 mRNA appeared linked through functional circuits, but receptor sites coincided with the presence of neurons co-expression of D2 and D3 mRNA suggests a functional positive for its mRNA. -

Characterization of Basal Forebrain Glutamate Neurons Suggests a Role in Control of Arousal and Avoidance Behavior
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.17.157479; this version posted June 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. This article is a US Government work. It is not subject to copyright under 17 USC 105 and is also made available for use under a CC0 license. Title: Characterization of basal forebrain glutamate neurons suggests a role in control of arousal and avoidance behavior Authors: James T. McKenna1, Chun Yang1, Thomas Bellio2, Marissa B. Anderson-Chernishof1, Mackenzie C. Gamble2, Abigail Hulverson2, John G. McCoy2, Stuart Winston1, James M. McNally1, Radhika Basheer1 and Ritchie E. Brown1 ORCIDs: James McKenna: 0000-0002-9710-3553 Chun Yang: 0000-0002-1979-0335 Radhika Basheer: 0000-0002-4052-6897 Ritchie Brown: 0000-0002-7164-4132 Institutional Affiliations: 1 Laboratory of Neuroscience, VA Boston Healthcare System and Harvard Medical School, Department of Psychiatry, 1400 VFW Parkway, West Roxbury, MA, 02132, USA. 2 Stonehill College, Easton, MA, 02357, USA. Running head: Basal Forebrain Glutamatergic Neurons Key words: calretinin, calbindin, parvalbumin, GAD67-GFP knock-in mice, nucleus basalis, ventral pallidum Corresponding author: Ritchie E. Brown, Dr. Rer. Nat., Address: Dept. of Psychiatry, VA Boston Healthcare System & Harvard Medical School, VA Medical Center, West Roxbury, 1400 VFW Parkway, MA, 02132, USA. Email: [email protected] Declarations: Funding: This work was supported by United States Veterans Administration Biomedical Laboratory Research and Development Service Merit Awards I01 BX004673, BX001356, I01 BX001404, I01 BX002774, I01 BX004500 and United States National Institute of Health support from NINDS R21 NS093000, NIMH R01 MH039683, NHLBI HL095491, R03- MH107650 and by SURE fellowships from Stonehill College.