Cyrus Chothia (1942–2019)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The SUPERFAMILY 2.0 Database: a Significant Proteome Update and a New Webserver Arun Prasad Pandurangan 1,*, Jonathan Stahlhacke2, Matt E
D490–D494 Nucleic Acids Research, 2019, Vol. 47, Database issue Published online 16 November 2018 doi: 10.1093/nar/gky1130 The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver Arun Prasad Pandurangan 1,*, Jonathan Stahlhacke2, Matt E. Oates2, Ben Smithers 2 and Julian Gough1 1MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK and 2Computer Science, University of Bristol, Bristol BS8 1UB, UK Received September 24, 2018; Revised October 23, 2018; Editorial Decision October 23, 2018; Accepted October 25, 2018 ABSTRACT level, most homologous proteins cluster together with high sequence similarity suggesting clear evolutionary relation- Here, we present a major update to the SUPERFAM- ship and functional consistency (3). The SUPERFAMILY ILY database and the webserver. We describe the ad- database provides domain annotations at both Superfamily dition of new SUPERFAMILY 2.0 profile HMM library and Family levels (4). containing a total of 27 623 HMMs. The database SUPERFAMILYprovides various analysis tools to facil- now includes Superfamily domain annotations for itate better analysis and interpretation of the database con- millions of protein sequences taken from the Uni- tent. They include the identification of under- and overrep- versal Protein Recourse Knowledgebase (UniPro- resentation of domains between genomes (5), construction tKB) and the National Center for Biotechnology In- of phylogenetic trees (6), analysis of the domain distribution formation (NCBI). This addition constitutes about 51 of superfamilies and families across the tree of life (7)aswell and 45 million distinct protein sequences obtained as providing ontology based annotations for SUPERFAM- ILY domains and architectures (8,9). -

Applied Category Theory for Genomics – an Initiative
Applied Category Theory for Genomics { An Initiative Yanying Wu1,2 1Centre for Neural Circuits and Behaviour, University of Oxford, UK 2Department of Physiology, Anatomy and Genetics, University of Oxford, UK 06 Sept, 2020 Abstract The ultimate secret of all lives on earth is hidden in their genomes { a totality of DNA sequences. We currently know the whole genome sequence of many organisms, while our understanding of the genome architecture on a systematic level remains rudimentary. Applied category theory opens a promising way to integrate the humongous amount of heterogeneous informations in genomics, to advance our knowledge regarding genome organization, and to provide us with a deep and holistic view of our own genomes. In this work we explain why applied category theory carries such a hope, and we move on to show how it could actually do so, albeit in baby steps. The manuscript intends to be readable to both mathematicians and biologists, therefore no prior knowledge is required from either side. arXiv:2009.02822v1 [q-bio.GN] 6 Sep 2020 1 Introduction DNA, the genetic material of all living beings on this planet, holds the secret of life. The complete set of DNA sequences in an organism constitutes its genome { the blueprint and instruction manual of that organism, be it a human or fly [1]. Therefore, genomics, which studies the contents and meaning of genomes, has been standing in the central stage of scientific research since its birth. The twentieth century witnessed three milestones of genomics research [1]. It began with the discovery of Mendel's laws of inheritance [2], sparked a climax in the middle with the reveal of DNA double helix structure [3], and ended with the accomplishment of a first draft of complete human genome sequences [4]. -

Functional Effects Detailed Research Plan
GeCIP Detailed Research Plan Form Background The Genomics England Clinical Interpretation Partnership (GeCIP) brings together researchers, clinicians and trainees from both academia and the NHS to analyse, refine and make new discoveries from the data from the 100,000 Genomes Project. The aims of the partnerships are: 1. To optimise: • clinical data and sample collection • clinical reporting • data validation and interpretation. 2. To improve understanding of the implications of genomic findings and improve the accuracy and reliability of information fed back to patients. To add to knowledge of the genetic basis of disease. 3. To provide a sustainable thriving training environment. The initial wave of GeCIP domains was announced in June 2015 following a first round of applications in January 2015. On the 18th June 2015 we invited the inaugurated GeCIP domains to develop more detailed research plans working closely with Genomics England. These will be used to ensure that the plans are complimentary and add real value across the GeCIP portfolio and address the aims and objectives of the 100,000 Genomes Project. They will be shared with the MRC, Wellcome Trust, NIHR and Cancer Research UK as existing members of the GeCIP Board to give advance warning and manage funding requests to maximise the funds available to each domain. However, formal applications will then be required to be submitted to individual funders. They will allow Genomics England to plan shared core analyses and the required research and computing infrastructure to support the proposed research. They will also form the basis of assessment by the Project’s Access Review Committee, to permit access to data. -

Meeting Review: Bioinformatics and Medicine – from Molecules To
Comparative and Functional Genomics Comp Funct Genom 2002; 3: 270–276. Published online 9 May 2002 in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/cfg.178 Feature Meeting Review: Bioinformatics And Medicine – From molecules to humans, virtual and real Hinxton Hall Conference Centre, Genome Campus, Hinxton, Cambridge, UK – April 5th–7th Roslin Russell* MRC UK HGMP Resource Centre, Genome Campus, Hinxton, Cambridge CB10 1SB, UK *Correspondence to: Abstract MRC UK HGMP Resource Centre, Genome Campus, The Industrialization Workshop Series aims to promote and discuss integration, automa- Hinxton, Cambridge CB10 1SB, tion, simulation, quality, availability and standards in the high-throughput life sciences. UK. The main issues addressed being the transformation of bioinformatics and bioinformatics- based drug design into a robust discipline in industry, the government, research institutes and academia. The latest workshop emphasized the influence of the post-genomic era on medicine and healthcare with reference to advanced biological systems modeling and simulation, protein structure research, protein-protein interactions, metabolism and physiology. Speakers included Michael Ashburner, Kenneth Buetow, Francois Cambien, Cyrus Chothia, Jean Garnier, Francois Iris, Matthias Mann, Maya Natarajan, Peter Murray-Rust, Richard Mushlin, Barry Robson, David Rubin, Kosta Steliou, John Todd, Janet Thornton, Pim van der Eijk, Michael Vieth and Richard Ward. Copyright # 2002 John Wiley & Sons, Ltd. Received: 22 April 2002 Keywords: bioinformatics; -

Tum1 Is Involved in the Metabolism of Sterol Esters in Saccharomyces Cerevisiae Katja Uršič1,4, Mojca Ogrizović1,Dušan Kordiš1, Klaus Natter2 and Uroš Petrovič1,3*
Uršič et al. BMC Microbiology (2017) 17:181 DOI 10.1186/s12866-017-1088-1 RESEARCHARTICLE Open Access Tum1 is involved in the metabolism of sterol esters in Saccharomyces cerevisiae Katja Uršič1,4, Mojca Ogrizović1,Dušan Kordiš1, Klaus Natter2 and Uroš Petrovič1,3* Abstract Background: The only hitherto known biological role of yeast Saccharomyces cerevisiae Tum1 protein is in the tRNA thiolation pathway. The mammalian homologue of the yeast TUM1 gene, the thiosulfate sulfurtransferase (a.k.a. rhodanese) Tst, has been proposed as an obesity-resistance and antidiabetic gene. To assess the role of Tum1 in cell metabolism and the putative functional connection between lipid metabolism and tRNA modification, we analysed evolutionary conservation of the rhodanese protein superfamily, investigated the role of Tum1 in lipid metabolism, and examined the phenotype of yeast strains expressing the mouse homologue of Tum1, TST. Results: We analysed evolutionary relationships in the rhodanese superfamily and established that its members are widespread in bacteria, archaea and in all major eukaryotic groups. We found that the amount of sterol esters was significantly higher in the deletion strain tum1Δ than in the wild-type strain. Expression of the mouse TST protein in the deletion strain did not rescue this phenotype. Moreover, although Tum1 deficiency in the thiolation pathway was complemented by re-introducing TUM1, it was not complemented by the introduction of the mouse homologue Tst. We further showed that the tRNA thiolation pathway is not involved in the regulation of sterol ester content in S. cerevisiae,asoverexpressionofthetEUUC,tKUUU and tQUUG tRNAs did not rescue the lipid phenotype in the tum1Δ deletion strain, and, additionally, deletion of the key gene for the tRNA thiolation pathway, UBA4, did not affect sterol ester content. -

Hmms Representing All Proteins of Known Structure. SCOP Sequence Searches, Alignments and Genome Assignments Julian Gough* and Cyrus Chothia
268–272 Nucleic Acids Research, 2002, Vol. 30, No. 1 © 2002 Oxford University Press SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments Julian Gough* and Cyrus Chothia MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK Received September 28, 2001; Revised and Accepted October 30, 2001 ABSTRACT (currently 59) covering approximately half of the The SUPERFAMILY database contains a library of soluble protein domains. The assignments, super- family breakdown and statistics on them are available hidden Markov models representing all proteins of from the server. The database is currently used by known structure. The database is based on the SCOP this group and others for genome annotation, structural ‘superfamily’ level of protein domain classification genomics, gene prediction and domain-based genomic which groups together the most distantly related studies. proteins which have a common evolutionary ancestor. There is a public server at http://supfam.org which provides three services: sequence searching, INTRODUCTION multiple alignments to sequences of known structure, The SUPERFAMILY database is based on the SCOP (1) and structural assignments to all complete genomes. classification of protein domains. SCOP is a structural domain- Given an amino acid or nucleotide query sequence based heirarchical classification with several levels including the server will return the domain architecture and the ‘superfamily’ level. Proteins grouped together at the super- SCOP classification. The server produces alignments family level are defined as having structural, functional and sequence evidence for a common evolutionary ancestor. It is at of the query sequences with sequences of known this level, as the name suggests, that SUPERFAMILY operates structure, and includes multiple alignments of because it is the level with the most distantly related protein genome and PDB sequences. -

Smurflite: Combining Simplified Markov Random Fields With
SMURFLite: combining simplified Markov random fields with simulated evolution improves remote homology detection for beta-structural proteins into the twilight zone Noah M. Daniels 1, Raghavendra Hosur 2, Bonnie Berger 2∗, and Lenore J. Cowen 1∗ 1Department of Computer Science, Tufts University, Medford, MA 02155 2Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139 ABSTRACT are limited in their power to recognize remote homologs because of Motivation: One of the most successful methods to date for their inability to model statistical dependencies between amino-acid recognizing protein sequences that are evolutionarily related has residues that are close in space but far apart in sequence (Lifson and been profile Hidden Markov Models (HMMs). However, these models Sander (1980); Zhu and Braun (1999); Olmea et al. (1999); Cowen do not capture pairwise statistical preferences of residues that are et al. (2002); Steward and Thorton (2002)). hydrogen bonded in beta sheets. These dependencies have been For this reason, many have suggested (White et al. (1994); partially captured in the HMM setting by simulated evolution in the Lathrop and Smith (1996); Thomas et al. (2008); Liu et al. (2009); training phase and can be fully captured by Markov Random Fields Menke et al. (2010); Peng and Xu (2011)) that more powerful (MRFs). However, the MRFs can be computationally prohibitive when Markov Random Fields (MRFs) be used. MRFs employ an auxiliary beta strands are interleaved in complex topologies. dependency graph which allows them to model more complex We introduce SMURFLite, a method that combines both simplified statistical dependencies, including statistical dependencies that Markov Random Fields and simulated evolution to substantially occur between amino-acid residues that are hydrogen bonded in beta improve remote homology detection for beta structures. -

AUTOPHY by Deepika Prasad a Thesis
ANALYZING MARKER GENE DIVERSITY USING AN AUTOMATED PHYLOGENETIC TOOL: AUTOPHY by Deepika Prasad A thesis submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics and Computational Biology Spring 2017 © 2017 Deepika Prasad All Rights Reserved ANALYZING MARKER GENE DIVERSITY USING AN AUTOMATED PHYLOGENETIC TOOL: AUTOPHY by Deepika Prasad Approved: __________________________________________________________ Shawn Polson, Ph.D. Professor in charge of thesis on behalf of the Advisory Committee Approved: __________________________________________________________ Kathleen F.McCoy, Ph.D. Chair of the Department of Computer and Information Sciences Approved: __________________________________________________________ Babatunde A. Ogunnaike, Ph.D. Dean of the College of Engineering Approved: __________________________________________________________ Ann L. Ardis, Ph.D. Senior Vice Provost for Graduate and Professional Education ACKNOWLEDGMENTS I want to express my sincerest gratitude to Professor Shawn Polson for his patience, continuous support, and enthusiasm. I could not have imagined having a better mentor to guide me through the process of my Master’s thesis. I would like to thank my committee members, Professor Eric Wommack and Honzhan Huang for their insightful comments and guidance in the thesis. I would also like to express my gratitude to Barbra Ferrell for asking important questions throughout my thesis research and helping me with the writing process. I would like to thank my friend Sagar Doshi, and lab mates Daniel Nasko, and Prasanna Joglekar, for helping me out with data, presentations, and giving important advice, whenever necessary. Last but not the least, I would like to thank my family, for being my pillars of strength. -
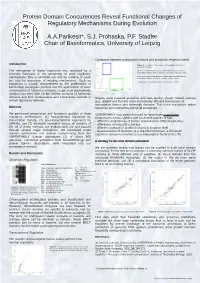
Protein Domain Coocurences Reveal Functional Changes of Regulatory Mechanisms During Evolution
Protein Domain Coocurences Reveal Functional Changes of Regulatory Mechanisms During Evolution A.A.Parikesit*, S.J. Prohaska, P.F. Stadler Chair of Bioinformatics, University of Leipzig Correlation between transcription factors and chromatin related proteins Introduction Figure 3 : Correlation of the number of transcription factors and chromatin domains. The emergence of higher organisms was facilitated by a Blue box. Species with few chromatin-related domains but many dramatic increases in the complexity of gene regulatory transcription factors: human, kangaroo rat, mouse, opossum, fugu. Green box: species with many chromatin-related domains but few mechanisms. This is achieved not only by addition of novel transcription factors: seq squirt, medaka, dolphin, yeast but also by expansion of existing mechanisms. Such an There are no organisms (known) that have a lot of both. expansion is usually characterized by the proliferation of functionally paralogous proteins and the appearance of novel combinations of functional domains. Large scale phylogenetic analysis can shed light on the relative amounts of functional domains and their combinations and interactions involved in Results show massive problems with data quality: closely related species certain regulatory networks. (e.g, dolphin and human) show dramatically different distributions of transcription factors and chromatin domains. This is not reasonable within Methods mammals and contradicts biological knowledge. We performed comparative and functional analysis of three SUPERFAMILY thus cannot be used for large-scale quantitative regulatory mechanisms: (1) transcriptional regulation by comparisons across species due to several sources of bias: transcription factors, (2) post-transcriptional regulation by - different completeness of protein annotation for different genomes miRNAs, and (3) chromatin regulation across all domains of - differences in transcript coverage life. -

Renaissance SUPERFAMILY in the Decade Since Its Launch, a Repository of Information About Proteins in Genomes Has Developed Into a Primary Reference
Renaissance SUPERFAMILY In the decade since its launch, a repository of information about proteins in genomes has developed into a primary reference. Dr Julian Gough, its creator, describes current work enhancing the scope, content and functionality of this key service Could you begin by outlining the reasons for How do HMMs feed into the service? creating SUPERFAMILY? HMMs are profi les which represent multiple SUPERFAMILY was originally created to better sequence alignments of homologous proteins understand molecular evolution, initially by in a rigorous statistical framework. They enabling comparison of the repertoire of proteins can be used to classify sequences based on and domains across the genomes of different homology and to create sequence alignments. species. The starting point was the Structural We use sequences of domains of known Classifi cation of Proteins (SCOP), and the most structure via iterative search procedures on basic purpose of SUPERFAMILY is to detect and large background sequence databases to classify these domains with known structural build alignments and, subsequently, models representatives in the protein sequences of representing domains of known structure at genomes. There are tens of thousands of known the superfamily level. These models are then protein structures, each the result of a costly searched against genome sequences to detect three-dimensional atomic resolution structure and classify the structural domains. determination by experiment, usually X-ray crystallography or nuclear magnetic resonance. -

An Expanded Evaluation of Protein Function Prediction Methods Shows an Improvement in Accuracy
The University of Southern Mississippi The Aquila Digital Community Faculty Publications 9-7-2016 An Expanded Evaluation of Protein Function Prediction Methods Shows an Improvement In Accuracy Yuxiang Jiang Indiana University TFollowal Ronnen this and Or onadditional works at: https://aquila.usm.edu/fac_pubs Buck P arInstitutet of the forGenomics Research Commons On Agin Wyatt T. Clark RecommendedYale University Citation AsmaJiang, YR.., OrBankapuron, T. R., Clark, W. T., Bankapur, A. R., D'Andrea, D., Lepore, R., Funk, C. S., Kahanda, I., Verspoor, K. M., Ben-Hur, A., Koo, D. E., Penfold-Brown, D., Shasha, D., Youngs, N., Bonneau, R., Lin, A., Sahraeian, S. Miami University M., Martelli, P. L., Profiti, G., Casadio, R., Cao, R., Zhong, Z., Cheng, J., Altenhoff, A., Skunca, N., Dessimoz, DanielC., Dogan, D'Andr T., Hakala,ea K., Kaewphan, S., Mehryar, F., Salakoski, T., Ginter, F., Fang, H., Smithers, B., Oates, M., UnivGough,ersity J., ofTör Romeönen, P., Koskinen, P., Holm, L., Chen, C., Hsu, W., Bryson, K., Cozzetto, D., Minneci, F., Jones, D. T., Chapan, S., BKC, D., Khan, I. K., Kihara, D., Ofer, D., Rappoport, N., Stern, A., Cibrian-Uhalte, E., Denny, P., Foulger, R. E., Hieta, R., Legge, D., Lovering, R. C., Magrane, M., Melidoni, A. N., Mutowo-Meullenet, P., SeePichler next, K., page Shypitsyna, for additional A., Li, B.,authors Zakeri, P., ElShal, S., Tranchevent, L., Das, S., Dawson, N. L., Lee, D., Lees, J. G., Stilltoe, I., Bhat, P., Nepusz, T., Romero, A. E., Sasidharan, R., Yang, H., Paccanaro, A., Gillis, J., Sedeño- Cortés, A. E., Pavlidis, P., Feng, S., Cejuela, J. -

Specialized Hidden Markov Model Databases for Microbial Genomics
Comparative and Functional Genomics Comp Funct Genom 2003; 4: 250–254. Published online 1 April 2003 in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/cfg.280 Conference Review Specialized hidden Markov model databases for microbial genomics Martin Gollery* University of Nevada, Reno, 1664 N. Virginia Street, Reno, NV 89557-0014, USA *Correspondence to: Abstract Martin Gollery, University of Nevada, Reno, 1664 N. Virginia As hidden Markov models (HMMs) become increasingly more important in the Street, Reno, NV analysis of biological sequences, so too have databases of HMMs expanded in size, 89557-0014, USA. number and importance. While the standard paradigm a short while ago was the E-mail: [email protected] analysis of one or a few sequences at a time, it has now become standard procedure to submit an entire microbial genome. In the future, it will be common to submit large groups of completed genomes to run simultaneously against a dozen public databases and any number of internally developed targets. This paper looks at some of the readily available HMM (or HMM-like) algorithms and several publicly available Received: 27 January 2003 HMM databases, and outlines methods by which the reader may develop custom Revised: 5 February 2003 HMM targets. Copyright 2003 John Wiley & Sons, Ltd. Accepted: 6 February 2003 Keywords: HMM; Pfam; InterPro; SuperFamily; TLfam; COG; TIGRfams Introduction will be a true homologue. As a result, HMMs have become very popular in the field of bioinformatics Over the last few years, hidden Markov models and a number of HMM databases have been (HMMs) have become one of the pre-eminent developed.