Rearrangements Within Human Spliceosomes Captured After Exon Ligation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-Like Mouse Models: Tracking the Role of the Hairless Gene
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2006 Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene Yutao Liu University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Liu, Yutao, "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino- like Mouse Models: Tracking the Role of the Hairless Gene. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1824 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yutao Liu entitled "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Doctor of Philosophy, with a major in Life Sciences. Brynn H. Voy, Major Professor We have read this dissertation and recommend its acceptance: Naima Moustaid-Moussa, Yisong Wang, Rogert Hettich Accepted for the Council: Carolyn R. -

Product Name: C9orf78 Polyclonal Antibody, ALEXA FLUOR® 647 Conjugated Catalog No
Product Name: C9orf78 Polyclonal Antibody, ALEXA FLUOR® 647 Conjugated Catalog No. : TAP01-84658R-A647 Intended Use: For Research Use Only. Not for used in diagnostic procedures. Size 100ul Concentration 1ug/ul Gene ID 51759 ISO Type Rabbit IgG Clone N/A Immunogen Range Conjugation ALEXA FLUOR® 647 Subcellular Locations Applications IF(IHC-P) Cross Reactive Species Human, Mouse, Rat Source KLH conjugated synthetic peptide derived from human C9orf78 Applications with IF(IHC-P)(1:50-200) Dilutions Purification Purified by Protein A. Background Chromosome 9 consists of about 145 million bases and 4% of the human genome and encodes nearly 900 genes. Considered to play a role in gender determination, deletion of the distal portion of 9p can lead to development of male to female sex reversal, the phenotype of a female with a male X,Y genotype. Hereditary hemorrhagic telangiectasia, which is characterized by harmful vascular defects, is associated with the chromosome 9 gene encoding endoglin protein, ENG. Familial dysautonomia is also associated with chromosome 9 though through the gene IKBKAP. Notably, chromosome 9 encompasses the largest interferon family gene cluster. Chromosome 9 is partnered with chromosome 22 in the translocation leading to the aberrant production of BCR-ABL fusion protein often found in leukemias. The C9orf78 gene product has been provisionally designated C9orf78 pending further characterization. Synonyms bA409K20.3; C9orf78; Chromosome 9 open reading frame 78; CI078_HUMAN; HCA59; Hepatocellular carcinoma associated antigen 59; Hepatocellular carcinoma-associated antigen 59; HSPC220; Uncharacterized protein C9orf78. Storage Aqueous buffered solution containing 1% BSA, 50% glycerol and 0.09% sodium azide. Store at 4°C for 12 months. -

Coupling of Spliceosome Complexity to Intron Diversity
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.19.436190; this version posted March 20, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Coupling of spliceosome complexity to intron diversity Jade Sales-Lee1, Daniela S. Perry1, Bradley A. Bowser2, Jolene K. Diedrich3, Beiduo Rao1, Irene Beusch1, John R. Yates III3, Scott W. Roy4,6, and Hiten D. Madhani1,6,7 1Dept. of Biochemistry and Biophysics University of California – San Francisco San Francisco, CA 94158 2Dept. of Molecular and Cellular Biology University of California - Merced Merced, CA 95343 3Department of Molecular Medicine The Scripps Research Institute, La Jolla, CA 92037 4Dept. of Biology San Francisco State University San Francisco, CA 94132 5Chan-Zuckerberg Biohub San Francisco, CA 94158 6Corresponding authors: [email protected], [email protected] 7Lead Contact 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.19.436190; this version posted March 20, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. SUMMARY We determined that over 40 spliceosomal proteins are conserved between many fungal species and humans but were lost during the evolution of S. cerevisiae, an intron-poor yeast with unusually rigid splicing signals. We analyzed null mutations in a subset of these factors, most of which had not been investigated previously, in the intron-rich yeast Cryptococcus neoformans. -

Parallel Molecular Evolution in Pathways, Genes, and Sites in High-Elevation Hummingbirds Revealed by Comparative Transcriptomics
GBE Parallel Molecular Evolution in Pathways, Genes, and Sites in High-Elevation Hummingbirds Revealed by Comparative Transcriptomics Marisa C.W. Lim1,*, Christopher C. Witt2, Catherine H. Graham1,3,andLilianaM.Davalos 1,4 1Department of Ecology and Evolution, Stony Brook University 2 Museum of Southwestern Biology and Department of Biology, University of New Mexico Downloaded from https://academic.oup.com/gbe/article-abstract/11/6/1552/5494706 by guest on 08 June 2019 3Swiss Federal Research Institute (WSL), Birmensdorf, Switzerland 4Consortium for Inter-Disciplinary Environmental Research, Stony Brook University *Corresponding author: E-mail: [email protected]. Accepted: May 12, 2019 Data deposition: The raw read data have been deposited in the NCBI Sequence Read Archive under BioProject: PRJNA543673, BioSample: SAMN11774663-SAMN11774674, SRA Study: SRP198856. All scripts used for analyses are available on Dryad: doi:10.5061/dryad.v961mb4. Abstract High-elevation organisms experience shared environmental challenges that include low oxygen availability, cold temperatures, and intense ultraviolet radiation. Consequently, repeated evolution of the same genetic mechanisms may occur across high-elevation taxa. To test this prediction, we investigated the extent to which the same biochemical pathways, genes, or sites were subject to parallel molecular evolution for 12 Andean hummingbird species (family: Trochilidae) representing several independent transitions to high elevation across the phylogeny. Across high-elevation species, we discovered parallel evolution for several pathways and genes with evidence of positive selection. In particular, positively selected genes were frequently part of cellular respiration, metabolism, or cell death pathways. To further examine the role of elevation in our analyses, we compared results for low- and high-elevation species and tested different thresholds for defining elevation categories. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
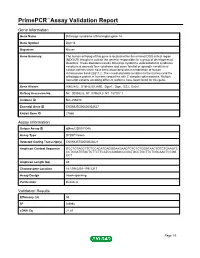
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DiGeorge syndrome critical region gene 14 Gene Symbol Dgcr14 Organism Mouse Gene Summary The human ortholog of this gene is located within the minimal DGS critical region (MDGCR) thought to contain the gene(s) responsible for a group of developmental disorders. These disorders include DiGeorge syndrome velocardiofacial syndrome conotruncal anomaly face syndrome and some familial or sporadic conotruncal cardiac defects which have been associated with microdeletion of human chromosome band 22q11.2. The encoded protein localizes to the nucleus and the orthologous protein in humans co-purifies with C complex spliceosomes. Multiple transcript variants encoding different isoforms have been found for this gene. Gene Aliases AI462402, D16H22S1269E, Dgcr1, Dgsi, ES2, Es2el RefSeq Accession No. NC_000082.6, NT_039624.8, NT_187007.1 UniGene ID Mm.256480 Ensembl Gene ID ENSMUSG00000003527 Entrez Gene ID 27886 Assay Information Unique Assay ID qMmuCID0011048 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000003621 Amplicon Context Sequence GCCTGTAGCTTCTCCACATCAGGGAAGAAGTCTCTCTGGATAACTGTCTGAAGTC CCTCGATGTACTCTTCTTCATCCAGGACCCGCTGCCTGCTTCTCGCAACTCCGG CCT Amplicon Length (bp) 82 Chromosome Location 16:17910201-17911217 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 95 R2 0.9994 cDNA Cq 21.87 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79.5 gDNA Cq 23.94 Specificity (%) 100 Information to assist with data interpretation is provided -

CRNKL1 Is a Highly Selective Regulator of Intron-Retaining HIV-1 and Cellular Mrnas
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 CRNKL1 is a highly selective regulator of intron-retaining HIV-1 and cellular mRNAs 2 3 4 Han Xiao1, Emanuel Wyler2#, Miha Milek2#, Bastian Grewe3, Philipp Kirchner4, Arif Ekici4, Ana Beatriz 5 Oliveira Villela Silva1, Doris Jungnickl1, Markus Landthaler2,5, Armin Ensser1, and Klaus Überla1* 6 7 1 Institute of Clinical and Molecular Virology, University Hospital Erlangen, Friedrich-Alexander 8 Universität Erlangen-Nürnberg, Erlangen, Germany 9 2 Berlin Institute for Medical Systems Biology, Max-Delbrück-Center for Molecular Medicine in the 10 Helmholtz Association, Robert-Rössle-Strasse 10, 13125, Berlin, Germany 11 3 Department of Molecular and Medical Virology, Ruhr-University, Bochum, Germany 12 4 Institute of Human Genetics, University Hospital Erlangen, Friedrich-Alexander Universität Erlangen- 13 Nürnberg, Erlangen, Germany 14 5 IRI Life Sciences, Institute für Biologie, Humboldt Universität zu Berlin, Philippstraße 13, 10115, Berlin, 15 Germany 16 # these two authors contributed equally 17 18 19 *Corresponding author: 20 Prof. Dr. Klaus Überla 21 Institute of Clinical and Molecular Virology, University Hospital Erlangen 22 Friedrich-Alexander Universität Erlangen-Nürnberg 23 Schlossgarten 4, 91054 Erlangen 24 Germany 25 Tel: (+49) 9131-8523563 26 e-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

PDF Datasheet
Product Datasheet Cactin Overexpression Lysate NBP2-06542 Unit Size: 0.1 mg Store at -80C. Avoid freeze-thaw cycles. Protocols, Publications, Related Products, Reviews, Research Tools and Images at: www.novusbio.com/NBP2-06542 Updated 3/17/2020 v.20.1 Earn rewards for product reviews and publications. Submit a publication at www.novusbio.com/publications Submit a review at www.novusbio.com/reviews/destination/NBP2-06542 Page 1 of 2 v.20.1 Updated 3/17/2020 NBP2-06542 Cactin Overexpression Lysate Product Information Unit Size 0.1 mg Concentration The exact concentration of the protein of interest cannot be determined for overexpression lysates. Please contact technical support for more information. Storage Store at -80C. Avoid freeze-thaw cycles. Buffer RIPA buffer Target Molecular Weight 88.5 kDa Product Description Description Transient overexpression lysate of chromosome 19 open reading frame 29 (C19orf29), transcript variant 2 The lysate was created in HEK293T cells, using Plasmid ID RC213573 and based on accession number NM_021231. The protein contains a C-MYC/DDK Tag. Gene ID 58509 Gene Symbol CACTIN Species Human Notes HEK293T cells in 10-cm dishes were transiently transfected with a non-lipid polymer transfection reagent specially designed and manufactured for large volume DNA transfection. Transfected cells were cultured for 48hrs before collection. The cells were lysed in modified RIPA buffer (25mM Tris-HCl pH7.6, 150mM NaCl, 1% NP-40, 1mM EDTA, 1xProteinase inhibitor cocktail mix, 1mM PMSF and 1mM Na3VO4, and then centrifuged to clarify the lysate. Protein concentration was measured by BCA protein assay kit.This product is manufactured by and sold under license from OriGene Technologies and its use is limited solely for research purposes. -

The Splicing Factor XAB2 Interacts with ERCC1-XPF and XPG for RNA-Loop Processing During Mammalian Development
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.20.211441; this version posted July 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Splicing Factor XAB2 interacts with ERCC1-XPF and XPG for RNA-loop processing during mammalian development Evi Goulielmaki1*, Maria Tsekrekou1,2*, Nikos Batsiotos1,2, Mariana Ascensão-Ferreira3, Eleftheria Ledaki1, Kalliopi Stratigi1, Georgia Chatzinikolaou1, Pantelis Topalis1, Theodore Kosteas1, Janine Altmüller4, Jeroen A. Demmers5, Nuno L. Barbosa-Morais3, George A. Garinis1,2* 1. Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology- Hellas, GR70013, Heraklion, Crete, Greece, 2. Department of Biology, University of Crete, Heraklion, Crete, Greece, 3. Instituto de Medicina Molecular João Lobo Antunes, Faculdade de Medicina da Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028 Lisboa, Portugal, 4. Cologne Center for Genomics (CCG), Institute for Genetics, University of Cologne, 50931, Cologne, Germany, 5. Proteomics Center, Netherlands Proteomics Center, and Department of Biochemistry, Erasmus University Medical Center, the Netherlands. Corresponding author: George A. Garinis ([email protected]) *: equally contributing authors bioRxiv preprint doi: https://doi.org/10.1101/2020.07.20.211441; this version posted July 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract RNA splicing, transcription and the DNA damage response are intriguingly linked in mammals but the underlying mechanisms remain poorly understood. Using an in vivo biotinylation tagging approach in mice, we show that the splicing factor XAB2 interacts with the core spliceosome and that it binds to spliceosomal U4 and U6 snRNAs and pre-mRNAs in developing livers. -

Visual Exploration Across Biomedical Databases
1 Visual Exploration Across Biomedical Databases Michael D. Lieberman∗†‡ Sima Taheri∗ Huimin Guo∗ Fatemeh Mir-Rashed∗ [email protected] [email protected] [email protected] [email protected] Inbal Yahav§ Aleks Aris∗ Ben Shneiderman∗† [email protected] [email protected] [email protected] Abstract—Though biomedical research often draws on knowl- mentioned in d. d may also have associations with other edge from a wide variety of fields, few visualization methods PubMed documents that cite d as a reference, as well as for biomedical data incorporate meaningful cross-database ex- associations to the PubMed documents that d itself cites. ploration. A new approach is offered for visualizing and explor- ∈ ing a query-based subset of multiple heterogeneous biomedical Furthermore, each gene g G could have associations with databases. Databases are modeled as an entity-relation graph the proteins for which g codes, or the DNA sequences in containing nodes (database records) and links (relationships which g’s code appears. Usually, the various types of records between records). Users specify a keyword search string to in these databases also have many attributes associated with retrieve an initial set of nodes, and then explore intra- and inter- them. For example, PubMed documents might be annotated database links. Results are visualized with user-defined semantic substrates to take advantage of the rich set of attributes usually with the date of publication, authors, and general topics, while present in biomedical data. Comments from domain experts gene records could be annotated with the relevant species, indicate that this visualization method is potentially advantageous location on chromosome, or function. -

Characterisation of Expression Patterns and Functional Role of Cactin in Early Zebrafish Development
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by MURAL - Maynooth University Research Archive Library Gene Expression Patterns 10 (2010) 199–206 Contents lists available at ScienceDirect Gene Expression Patterns journal homepage: www.elsevier.com/locate/gep Characterisation of expression patterns and functional role of Cactin in early zebrafish development Paola Atzei a, Fan Yang b, Ross Collery b, Breandan N. Kennedy b,1, Paul N. Moynagh a,*,1 a Institute of Immunology, National University of Ireland Maynooth, Maynooth, Co. Kildare, Ireland b UCD School of Biomolecular and Biomedical Sciences, UCD Conway Institute, University College Dublin, Dublin 4, Ireland article info abstract Article history: The immune system of teleost zebrafish (Danio rerio) shows high similarity to mammalian counterparts Received 21 December 2009 sharing many innate immune components including Toll-Like Receptors (TLRs), cytokines, chemokines Received in revised form 12 March 2010 and complement molecules. As in mammals, zebrafish also contains the transcription factor NF-jB that Accepted 19 March 2010 plays dualist roles in innate immunity and early development. Indeed NF-jB members are expressed in Available online 27 March 2010 different temporal patterns during the early stages of zebrafish embryogenesis indicating that each mol- ecule is involved in specific developmental events. In the present study we employ zebrafish as a model Keywords: to characterise the expression pattern and role of a novel NF-jB regulator, termed Cactin, in early devel- Toll-like receptors opment. Cactin was first characterised in Drosophila as a new member of the Rel pathway that could NF-jB Cactin affect the generation of dorsal–ventral polarity. -

Molecular Effects of Isoflavone Supplementation Human Intervention Studies and Quantitative Models for Risk Assessment
Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis committee Promotors Prof. Dr Pieter van ‘t Veer Professor of Nutritional Epidemiology Wageningen University Prof. Dr Evert G. Schouten Emeritus Professor of Epidemiology and Prevention Wageningen University Co-promotors Dr Anouk Geelen Assistant professor, Division of Human Nutrition Wageningen University Dr Lydia A. Afman Assistant professor, Division of Human Nutrition Wageningen University Other members Prof. Dr Jaap Keijer, Wageningen University Dr Hubert P.J.M. Noteborn, Netherlands Food en Consumer Product Safety Authority Prof. Dr Yvonne T. van der Schouw, UMC Utrecht Dr Wendy L. Hall, King’s College London This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Molecular effects of isoflavone supplementation Human intervention studies and quantitative models for risk assessment Vera van der Velpen Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 20 June 2014 at 13.30 p.m. in the Aula. Vera van der Velpen Molecular effects of isoflavone supplementation: Human intervention studies and quantitative models for risk assessment 154 pages PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN: 978-94-6173-952-0 ABSTRact Background: Risk assessment can potentially be improved by closely linked experiments in the disciplines of epidemiology and toxicology.