Phil. Trans. R. Soc. B - Issue
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LARVAL FORMS in ECHINODERMATA in Echinoderms
LARVAL FORMS IN ECHINODERMATA In echinoderms eggs and sperms are released in water and fertilization takes place in water forming zygote. Echinoderms are deuterostomes and hence cleavage is radial, holoblastic and indeterminate. The larvae hatch in water and feed and grow through successive larval stages to become adults. The larvae of echinoderms are bilaterally symmetrical but lose symmetry during metamorphosis. Different classes of echinoderms show structurally different larval stages and their comparisons can reveal their evolutionary ancestry. LARVAE OF ASTEROIDEA There are three larval stages in Asteroidea in the course of their development to adult stage. Early bipinnaria appears like hypothetical dipleurula. It has oval body without arms and ciliary bands for locomotion. It has well developed alimentary canal for feeding and grows to become bipinnaria. Bipinnaria larva possesses 5 pairs of ciliated arms which do not have any skeletal support inside. These arms are used for swimming in water while feeding on planktons. Preoral and postoral ciliary bands are also present. This larva resembles auricularia larva of Holothuroidea in general appearance. Brachiolaria larva is formed after 6-7 weeks of life and growth of bipinnaria. This larva is sedentary and remains attached to a hard substratum for which it possesses three brachiolarian arms having adhesive discs at the tip. Ciliated arms get reduced and become thin and functionless, while mouth, anus and gut are well developed. It has axocoel, hydocoel and somatocoel that later on give rise to water vascular system Development of starfish takes place inside the sedentary brachiolaria which ruptures and releases tiny starfish into water. LARVAE OF HOLOTHUROIDEA Class Holothuroidea demonstrate two larval stages, namely, auricularia and doliolaria larvae. -

A New Record of the Persian Brook Salamander, Paradactylodon Persicus (Eiselt & Steiner, 1970) (Amphibia: Caudata: Hynobiidae) in Northern Iran
Bonn zoological Bulletin Volume 60 Issue 1 pp. 63–65 Bonn, May 2011 A new record of the Persian Brook Salamander, Paradactylodon persicus (Eiselt & Steiner, 1970) (Amphibia: Caudata: Hynobiidae) in northern Iran Faraham Ahmadzadeh1, 2*, Fatemeh Khanjani3, Aref Shadkam4 & Wolfgang Böhme2 1Department of Biodiversity and Ecosystem Management, Environmental Sciences Research Institute, Shahid Beheshti University, Evin, Tehran, G. C., Iran 2Herpetology Section, Zoologisches Forschungsmuseum Alexander Koenig (ZFMK), Adenauerallee 160, D-53113 Bonn, Germany 3Department of Habitat and Biodiversity, Faculty of Environment and Energy, Science and Research Branch, Islamic Azad University, Ponak, Tehran, Iran 4Department of Environment, Rezvan Shahr, Province Gilan, Iran *Corresponding Email address: [email protected]. INTRODUCTION The Persian Brook Salamander, Paradactylodon persicus 90 mm; tail length 120 mm; head large, 20 mm in length; (Eiselt & Steiner, 1970) is an endemic and poorly known vomerine teeth in two arch-shaped rows; snout rounded; species of northern Iran (Baloutch & Kami 1995; Kami fore and hind limbs with four digits; tail flattened later- 1999). It was originally described as Batrachuperus per- ally, with round-tapered end; dorsal head and body, as well sicus by Eiselt & Steiner (1970), but has been transferred as upper surface of tail brownish with yellow spots and to the genus Paradactylodon based on genetic studies by marblings; belly cream without pattern (Fig. 2a–b). Zhang et al. (2006). This species has been reported from two localities only: Weyser, southeast of Chalus, in Paradactylodon persicus inhabits the mountainous Mazandaran Province (36º 30´ 35” N and 51º 26´ 38” E) streams and brooks, with cool, fast-flowing water and Delmadeh village, southeast of Khalkhal, in Ardabil (Baloutch & Kami 1995; Kami 1999; Ahmadzadeh & Ka- Province (37º 22´ 34” N and 48º 47´ 35” E) (Kami 2004; mi 2009). -

1. Project Title: an Ex-Situ Initiative to Rescue Merida's Whistling Frog, An
1. Project Title: An ex-situ initiative to rescue Merida’s Whistling Frog, an endangered undescribed Leptodactylus species. 2. Names, institutional affiliation, and email address of project leader: Enrique La Marca, Laboratory of Biogeography of the University of Los Andes at Merida, Venezuela. E-mail: [email protected] Member of the IUCN SSC Amphibian Specialist Group of Experts, since 1995. 3. Total funding amount requested from Amphibian Ark in USD$: US$ 5.000,oo 4. Executive summary (300 words or less) This project is aimed to rescue populations of the Merida’s Whistling Frog, an endangered undescribed Leptodactylus species. This is a Venezuelan amphibian with a much-restricted distribution in a highly perturbed urban habitat. The species has not been described yet, but is already known to be in peril. This will be the second Venezuelan Leptodactylus in need of protection besides Leptodactylus magistris , an endemic species living outside the Andes (Mijares and La Marca 2004, La Marca, Mijares and Señaris 2008, La Marca et al . 2013). No special measurements have been taken to date to protect those species, nor there is official protection for the habitat where they lives. The aim of this project it to set an ex-situ conservation program for the Merida’s Whistling Frog and to produce husbandry guidelines for the same that could be useful for other such Leptodactylus species in risk. There is an urgent need for conservation of this species of Leptodactylus . It shares the same geographic distribution Mannophryne collaris (although not the same habitat requirements; i.e. sympatran but not syntopic species). -

Amphibiaweb's Illustrated Amphibians of the Earth
AmphibiaWeb's Illustrated Amphibians of the Earth Created and Illustrated by the 2020-2021 AmphibiaWeb URAP Team: Alice Drozd, Arjun Mehta, Ash Reining, Kira Wiesinger, and Ann T. Chang This introduction to amphibians was written by University of California, Berkeley AmphibiaWeb Undergraduate Research Apprentices for people who love amphibians. Thank you to the many AmphibiaWeb apprentices over the last 21 years for their efforts. Edited by members of the AmphibiaWeb Steering Committee CC BY-NC-SA 2 Dedicated in loving memory of David B. Wake Founding Director of AmphibiaWeb (8 June 1936 - 29 April 2021) Dave Wake was a dedicated amphibian biologist who mentored and educated countless people. With the launch of AmphibiaWeb in 2000, Dave sought to bring the conservation science and basic fact-based biology of all amphibians to a single place where everyone could access the information freely. Until his last day, David remained a tirelessly dedicated scientist and ally of the amphibians of the world. 3 Table of Contents What are Amphibians? Their Characteristics ...................................................................................... 7 Orders of Amphibians.................................................................................... 7 Where are Amphibians? Where are Amphibians? ............................................................................... 9 What are Bioregions? ..................................................................................10 Conservation of Amphibians Why Save Amphibians? ............................................................................. -

Testis Development and Differentiation in Amphibians
G C A T T A C G G C A T genes Review Testis Development and Differentiation in Amphibians Álvaro S. Roco , Adrián Ruiz-García and Mónica Bullejos * Departamento de Biología Experimental, Facultad de Ciencias Experimentales, Campus Las Lagunillas S/N, Universidad de Jaén, 23071 Jaén, Spain; [email protected] (Á.S.R.); [email protected] (A.R.-G.) * Correspondence: [email protected]; Tel.: +34-953-212770 Abstract: Sex is determined genetically in amphibians; however, little is known about the sex chromosomes, testis-determining genes, and the genes involved in testis differentiation in this class. Certain inherent characteristics of the species of this group, like the homomorphic sex chromosomes, the high diversity of the sex-determining mechanisms, or the existence of polyploids, may hinder the design of experiments when studying how the gonads can differentiate. Even so, other features, like their external development or the possibility of inducing sex reversal by external treatments, can be helpful. This review summarizes the current knowledge on amphibian sex determination, gonadal development, and testis differentiation. The analysis of this information, compared with the information available for other vertebrate groups, allows us to identify the evolutionarily conserved and divergent pathways involved in testis differentiation. Overall, the data confirm the previous observations in other vertebrates—the morphology of the adult testis is similar across different groups; however, the male-determining signal and the genetic networks involved in testis differentiation are not evolutionarily conserved. Keywords: amphibian; sex determination; gonadal differentiation; testis; sex reversal Citation: Roco, Á.S.; Ruiz-García, A.; Bullejos, M. Testis Development and Differentiation in Amphibians. -
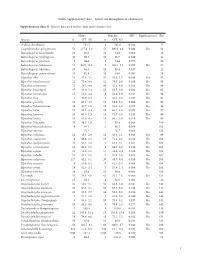
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

AMPHIBIA: ANURA: LEPTODACTYLIDAE Leptodactylus Pentadactylus
887.1 AMPHIBIA: ANURA: LEPTODACTYLIDAE Leptodactylus pentadactylus Catalogue of American Amphibians and Reptiles. Heyer, M.M., W.R. Heyer, and R.O. de Sá. 2011. Leptodactylus pentadactylus . Leptodactylus pentadactylus (Laurenti) Smoky Jungle Frog Rana pentadactyla Laurenti 1768:32. Type-locality, “Indiis,” corrected to Suriname by Müller (1927: 276). Neotype, Nationaal Natuurhistorisch Mu- seum (RMNH) 29559, adult male, collector and date of collection unknown (examined by WRH). Rana gigas Spix 1824:25. Type-locality, “in locis palu - FIGURE 1. Leptodactylus pentadactylus , Brazil, Pará, Cacho- dosis fluminis Amazonum [Brazil]”. Holotype, Zoo- eira Juruá. Photograph courtesy of Laurie J. Vitt. logisches Sammlung des Bayerischen Staates (ZSM) 89/1921, now destroyed (Hoogmoed and Gruber 1983). See Nomenclatural History . Pre- lacustribus fluvii Amazonum [Brazil]”. Holotype, occupied by Rana gigas Wallbaum 1784 (= Rhin- ZSM 2502/0, now destroyed (Hoogmoed and ella marina {Linnaeus 1758}). Gruber 1983). Rana coriacea Spix 1824:29. Type-locality: “aquis Rana pachypus bilineata Mayer 1835:24. Type-local MAP . Distribution of Leptodactylus pentadactylus . The locality of the neotype is indicated by an open circle. A dot may rep - resent more than one site. Predicted distribution (dark-shaded) is modified from a BIOCLIM analysis. Published locality data used to generate the map should be considered as secondary sources, as we did not confirm identifications for all specimen localities. The locality coordinate data and sources are available on a spread sheet at http://learning.richmond.edu/ Leptodactylus. 887.2 FIGURE 2. Tadpole of Leptodactylus pentadactylus , USNM 576263, Brazil, Amazonas, Reserva Ducke. Scale bar = 5 mm. Type -locality, “Roque, Peru [06 o24’S, 76 o48’W].” Lectotype, Naturhistoriska Riksmuseet (NHMG) 497, age, sex, collector and date of collection un- known (not examined by authors). -

Bonn Zoological Bulletin Volume 60 Issue 1 Pp
1 © Biodiversity Heritage Library, http://www.biodiversitylibrary.org/; www.zoologicalbulletin.de; www.biologiezentrum.at Bonn zoological Bulletin Volume 60 Issue 1 pp. 63-65 Bonn, May 201 A new record of the Persian Brook Salamander, Paradactylodon persicus (Eiselt & Steiner, 1970) (Amphibia: Caudata: Hynobiidae) in northern Iran 1 2 3 4 2 - Faraham Ahmadzadeh \ Fatemeh Khanjani , Aref Shadkam & Wolfgang Bohme 'Department of Biodiversity and Ecosystem Management, Environmental Sciences Research Institute, Shahid Beheshti University, Evin, Tehran, G. C, Iran 2 Herpetology Section, Zoologisches Forschungsmuseum Alexander Koenig (ZFMK), Adenauerallee 160, D-53113 Bonn, Germany 3 Department of Habitat and Biodiversity, Faculty of Environment and Energy, Science and Research Branch, Islamic Azad University, Ponak, Tehran, Iran 4 Department of Environment, Rezvan Shahr, Province Gilan, Iran "Corresponding Email address: [email protected]. INTRODUCTION The Persian Brook Salamander, Paradactylodon persicus 90 mm; tail length 120 mm; head large, 20 mm in length; (Eiselt & Steiner, 1970) is an endemic and poorly known vomerine teeth in two arch-shaped rows; snout rounded; species of northern Iran (Baloutch & Kami 1995; Kami fore and hind limbs with four digits; tail flattened later- 1999). It was originally described as Batrachuperus per- ally, with round-tapered end; dorsal head and body, as well sicus by Eiselt & Steiner ( 1 970), but has been transferred as upper surface of tail brownish with yellow spots and to the genus Paradactylodon based on genetic studies by marblings; belly cream without pattern (Fig. 2a-b). Zhang et al. (2006). This species has been reported from two localities only: Weyser, southeast of Chalus, in Paradactylodon persicus inhabits the mountainous Mazandaran Province (36° 30' 35" N and 51° 26' 38" E) streams and brooks, with cool, fast-flowing water and Delmadeh village, southeast of Khalkhal, in Ardabil (Baloutch & Kami 1995; Kami 1999; Ahmadzadeh & Ka- Province (37° 22' 34" N and 48° 47' 35" E) (Kami 2004; mi 2009). -

©Copyright 2008 Joseph A. Ross the Evolution of Sex-Chromosome Systems in Stickleback Fishes
©Copyright 2008 Joseph A. Ross The Evolution of Sex-Chromosome Systems in Stickleback Fishes Joseph A. Ross A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2008 Program Authorized to Offer Degree: Molecular and Cellular Biology University of Washington Graduate School This is to certify that I have examined this copy of a doctoral dissertation by Joseph A. Ross and have found that it is complete and satisfactory in all respects, and that any and all revisions required by the final examining committee have been made. Chair of the Supervisory Committee: Catherine L. Peichel Reading Committee: Catherine L. Peichel Steven Henikoff Barbara J. Trask Date: In presenting this dissertation in partial fulfillment of the requirements for the doctoral degree at the University of Washington, I agree that the Library shall make its copies freely available for inspection. I further agree that extensive copying of the dissertation is allowable only for scholarly purposes, consistent with “fair use” as prescribed in the U.S. Copyright Law. Requests for copying or reproduction of this dissertation may be referred to ProQuest Information and Learning, 300 North Zeeb Road, Ann Arbor, MI 48106-1346, 1-800-521-0600, to whom the author has granted “the right to reproduce and sell (a) copies of the manuscript in microform and/or (b) printed copies of the manuscript made from microform.” Signature Date University of Washington Abstract The Evolution of Sex-Chromosome Systems in Stickleback Fishes Joseph A. Ross Chair of the Supervisory Committee: Affiliate Assistant Professor Catherine L. -

3Systematics and Diversity of Extant Amphibians
Systematics and Diversity of 3 Extant Amphibians he three extant lissamphibian lineages (hereafter amples of classic systematics papers. We present widely referred to by the more common term amphibians) used common names of groups in addition to scientifi c Tare descendants of a common ancestor that lived names, noting also that herpetologists colloquially refer during (or soon after) the Late Carboniferous. Since the to most clades by their scientifi c name (e.g., ranids, am- three lineages diverged, each has evolved unique fea- bystomatids, typhlonectids). tures that defi ne the group; however, salamanders, frogs, A total of 7,303 species of amphibians are recognized and caecelians also share many traits that are evidence and new species—primarily tropical frogs and salaman- of their common ancestry. Two of the most defi nitive of ders—continue to be described. Frogs are far more di- these traits are: verse than salamanders and caecelians combined; more than 6,400 (~88%) of extant amphibian species are frogs, 1. Nearly all amphibians have complex life histories. almost 25% of which have been described in the past Most species undergo metamorphosis from an 15 years. Salamanders comprise more than 660 species, aquatic larva to a terrestrial adult, and even spe- and there are 200 species of caecilians. Amphibian diver- cies that lay terrestrial eggs require moist nest sity is not evenly distributed within families. For example, sites to prevent desiccation. Thus, regardless of more than 65% of extant salamanders are in the family the habitat of the adult, all species of amphibians Plethodontidae, and more than 50% of all frogs are in just are fundamentally tied to water. -

Download the PDF Article
DIRECTEUR DE LA PUBLICATION : Bruno David Président du Muséum national d’Histoire naturelle RÉDACTRICE EN CHEF / EDITOR-IN-CHIEF : Laure Desutter-Grandcolas ASSISTANTS DE RÉDACTION / ASSISTANT EDITORS : Anne Mabille ([email protected]), Emmanuel Côtez MISE EN PAGE / PAGE LAYOUT : Anne Mabille COMITÉ SCIENTIFIQUE / SCIENTIFIC BOARD : James Carpenter (AMNH, New York, États-Unis) Maria Marta Cigliano (Museo de La Plata, La Plata, Argentine) Henrik Enghoff (NHMD, Copenhague, Danemark) Rafael Marquez (CSIC, Madrid, Espagne) Peter Ng (University of Singapore) Norman I. Platnick (AMNH, New York, États-Unis) Jean-Yves Rasplus (INRA, Montferrier-sur-Lez, France) Jean-François Silvain (IRD, Gif-sur-Yvette, France) Wanda M. Weiner (Polish Academy of Sciences, Cracovie, Pologne) John Wenzel (The Ohio State University, Columbus, États-Unis) COUVERTURE / COVER : Podoces pleskei Zarudny, 1896. Photo by M. Ghorbani. Zoosystema est indexé dans / Zoosystema is indexed in: – Science Citation Index Expanded (SciSearch®) – ISI Alerting Services® – Current Contents® / Agriculture, Biology, and Environmental Sciences® – Scopus® Zoosystema est distribué en version électronique par / Zoosystema is distributed electronically by: – BioOne® (http://www.bioone.org) Les articles ainsi que les nouveautés nomenclaturales publiés dans Zoosystema sont référencés par / Articles and nomenclatural novelties published in Zoosystema are referenced by: – ZooBank® (http://zoobank.org) Zoosystema est une revue en fl ux continu publiée par les Publications scientifi ques du Muséum, Paris / Zoosystema is a fast track journal published by the Museum Science Press, Paris Les Publications scientifi ques du Muséum publient aussi / The Museum Science Press also publish: Adansonia, Anthropozoologica, European Journal of Taxonomy, Geodiversitas, Naturae. Diff usion – Publications scientifi ques Muséum national d’Histoire naturelle CP 41 – 57 rue Cuvier F-75231 Paris cedex 05 (France) Tél. -

Predatory Ecology of the Invasive Wrinkled Frog (Glandirana Rugosa) in Hawai´I
Gut check: predatory ecology of the invasive wrinkled frog (Glandirana rugosa) in Hawai´i By Melissa J. Van Kleeck and Brenden S. Holland* Abstract Invertebrates constitute the most diverse Pacific island animal lineages, and have correspondingly suffered the most significant extinction rates. Losses of native invertebrate lineages have been driven largely by ecosystem changes brought about by loss of habitat and direct predation by introduced species. Although Hawaii notably lacks native terrestrial reptiles and amphibians, both intentional and unintentional anthropogenic releases of herpetofauna have resulted in the establishment of more than two dozen species of frogs, toads, turtles, lizards, and a snake. Despite well-known presence of nonnative predatory species in Hawaii, ecological impacts remain unstudied for a majority of these species. In this study, we evaluated the diet of the Japanese wrinkled frog, Glandirana rugosa, an intentional biocontrol release in the Hawaiian Islands in the late 19th century. We collected live frogs on Oahu and used museum collections from both Oahu and Maui to determine exploited diet composition. These data were then compared to a published dietary analysis from the native range in Japan. We compiled and summarized field and museum distribution data from Oahu, Maui, and Kauai to document the current range of this species. Gut content analyses suggest that diet composition in the Hawaiian Islands is significantly different from that that in its native Japan. In the native range, the dominant taxonomic groups by volume were Coleoptera (beetles), Lepidoptera (moths, butterflies) and Formicidae (ants). Invasive frogs in Hawaii exploited mostly Dermaptera (earwigs), Amphipoda (landhoppers) and Hemiptera (true bugs).