Fine-Tuning the Chromosome Ends the Last Base of Human Telomeres
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Introduction of Human Telomerase Reverse Transcriptase to Normal Human Fibroblasts Enhances DNA Repair Capacity
Vol. 10, 2551–2560, April 1, 2004 Clinical Cancer Research 2551 Introduction of Human Telomerase Reverse Transcriptase to Normal Human Fibroblasts Enhances DNA Repair Capacity Ki-Hyuk Shin,1 Mo K. Kang,1 Erica Dicterow,1 INTRODUCTION Ayako Kameta,1 Marcel A. Baluda,1 and Telomerase, which consists of the catalytic protein subunit, No-Hee Park1,2 human telomerase reverse transcriptase (hTERT), the RNA component of telomerase (hTR), and several associated pro- 1School of Dentistry and 2Jonsson Comprehensive Cancer Center, University of California, Los Angeles, California teins, has been primarily associated with maintaining the integ- rity of cellular DNA telomeres in normal cells (1, 2). Telomer- ase activity is correlated with the expression of hTERT, but not ABSTRACT with that of hTR (3, 4). Purpose: From numerous reports on proteins involved The involvement of DNA repair proteins in telomere main- in DNA repair and telomere maintenance that physically tenance has been well documented (5–8). In eukaryotic cells, associate with human telomerase reverse transcriptase nonhomologous end-joining requires a DNA ligase and the (hTERT), we inferred that hTERT/telomerase might play a DNA-activated protein kinase, which is recruited to the DNA role in DNA repair. We investigated this possibility in nor- ends by the DNA-binding protein Ku. Ku binds to hTERT mal human oral fibroblasts (NHOF) with and without ec- without the need for telomeric DNA or hTR (9), binds the topic expression of hTERT/telomerase. telomere repeat-binding proteins TRF1 (10) and TRF2 (11), and Experimental Design: To study the effect of hTERT/ is thought to regulate the access of telomerase to telomere DNA telomerase on DNA repair, we examined the mutation fre- ends (12, 13). -

T-Loops and the Origin of Telomeres E
PERSPECTIVES 66. Steinman, R. M., Mellman, I. S., Muller, W. A. & Cohn, Z. A. Acknowledgments eukaryotes evolved. The presence of t-loops at Endocytosis and the recycling of plasma membrane. H.S. is supported by the Norwegian Cancer Society and the J. Cell Biol. 96, 1–27 (1983). Research Council of Norway, and J.G. by the Swiss National present-day telomeres and their association 67. Lafont, F., Lecat, S., Verkade, P. & Simons, K. Annexin Science Foundation and the Human Frontier Science Programme with proteins that have evolved from RDR XIIIb associates with lipid microdomains to function in Organization. apical delivery. J. Cell Biol. 142, 1413–1427 (1998). factors might be remnants of the original 68. Aniento, F., Gu, F., Parton, R. & Gruenberg, J. Competing interests statement telomere system. Furthermore, the relative An endosomal βcop is involved in the pH-dependent The authors declare that they have no competing financial interests. formation of transport vesicles destined for late ease with which many eukaryotes can main- endosomes. J. Cell Biol. 133, 29–41 (1996). tain telomeres without telomerase might 69. Gu, F., Aniento, F., Parton, R. & Gruenberg, J. Functional Online links dissection of COP-I subunits in the biogenesis of reflect this ancient system of chromosome-end multivesicular endosomes. J. Cell Biol. 139, 1183–1195 DATABASES replication. This proposal ends with a dis- (1997). The following terms in this article are linked online to: 70. Gu, F. & Gruenberg, J. ARF1 regulates pH-dependent Interpro: http://www.ebi.ac.uk/interpro/ cussion of the advantages of the telom- COP functions in the early endocytic pathway. -

Telomeres.Pdf
Telomeres Secondary article Elizabeth H Blackburn, University of California, San Francisco, California, USA Article Contents . Introduction Telomeres are specialized DNA–protein structures that occur at the ends of eukaryotic . The Replication Paradox chromosomes. A special ribonucleoprotein enzyme called telomerase is required for the . Structure of Telomeres synthesis and maintenance of telomeric DNA. Synthesis of Telomeric DNA by Telomerase . Functions of Telomeres Introduction . Telomere Homeostasis . Alternatives to Telomerase-generated Telomeric DNA Telomeres are the specialized chromosomal DNA–protein . Evolution of Telomeres and Telomerase structures that comprise the terminal regions of eukaryotic chromosomes. As discovered through studies of maize and somes. One critical part of this protective function is to fruitfly chromosomes in the 1930s, they are required to provide a means by which the linear chromosomal DNA protect and stabilize the genetic material carried by can be replicated completely, without the loss of terminal eukaryotic chromosomes. Telomeres are dynamic struc- DNA nucleotides from the 5’ end of each strand of this tures, with their terminal DNA being constantly built up DNA. This is necessary to prevent progressive loss of and degraded as dividing cells replicate their chromo- terminal DNA sequences in successive cycles of chromo- somes. One strand of the telomeric DNA is synthesized by somal replication. a specialized ribonucleoprotein reverse transcriptase called telomerase. Telomerase is required for both -

DNA Replication
9 DNA Replication To understand the chemistry Living cells must be able to duplicate their entire set of genetic instructions Goal of DNA synthesis and how every time they divide. Likewise, multicellular organisms must be able to DNA is replicated with high pass on complete copies of their genetic information to future generations. accuracy. This requires that the instructions are stored in a form that is capable of Objectives being duplicated. As we have seen (and will return to in Chapter 11), genetic instructions are embedded in the order of nucleobases in the DNA. As we After this chapter, you should be able to have also seen, the self-complementary nature of the double helix provides • describe the experiment that proved a simple (in principle) templating mechanism for replicating DNA into two that DNA replication is semi- identical copies. Only DNA (and in some instances RNA when, as in the conservative. case of the genomes of RNA viruses, it is used as a repository of genetic • diagram the reaction for information) are self-complementary and hence capable of being replicated. phosphodiester bond formation. The other three categories of macromolecules—carbohydrates, lipids, and • explain the energetics of DNA proteins—are not self-complementary and do not serve as templates for synthesis. their own production. Instead, the synthesis of these macromolecules is • explain why the 5’-to-3’ rule creates a ultimately directed by information stored in DNA through the action of conundrum during replication. enzymes and other proteins. Here we focus on the chemical and enzymatic • explain how DNA is replicated mechanisms by which DNA acts as a template for its own duplication and accurately. -

Getting Ready for DNA Duplication
INSIGHT INTRACELLULAR ORGANIZATION Getting ready for DNA duplication The discovery of a biomolecular condensate involved in DNA replication has wide-ranging implications. NINA Y YAO AND MICHAEL E O’DONNELL involves regions with different physical proper- Related research article Parker MW, Bell ties separating from each other). The molecules M, Mir M, Kao JA, Darzacq X, Botchan MR, required for the formation of condensates are Berger JM. 2019. A new class of disordered called scaffolding factors, while the molecules elements controls DNA replication through encapsulated inside are known as clients. A initiator self-assembly. eLife 8:e48562. DOI: given condensate typically contains only those 10.7554/eLife.48562 clients involved in the relevant reaction. Now, in eLife, Michael Botchan (UC Berkeley), James Berger (Johns Hopkins) and colleagues – including Matthew Parker as first author – report on the discovery of a biomolecular condensate esearch into biomolecular condensates – required for the initiation of DNA replication in self-contained regions within cells where the fruit fly Drosophila melanogaster R specific reactions take place – is taking (Parker et al., 2019). Three proteins – ORC, cell biology by storm. Biomolecular condensates Cdc6 and Cdt1 – form the scaffold along with do not have membranes, but they are able to DNA, while a hexamer ring protein called keep the proteins and/or nucleic acids involved Mcm2-7 is the client. During the G1 phase of the in a particular reaction separate from the rest of cell cycle, the scaffold proteins load two Mcm2- the cell. Biomolecular condensates do not have 7 hexamers onto the DNA to form the pre-repli- membranes, but they are able to keep the mole- cative complex, which marks the spot where cules involved in a particular reaction (usually DNA replication will begin (Figure 1A–C; proteins, but sometimes also nucleic acids) sepa- Bell and Labib, 2016; Bleichert et al., 2017). -

Telomere and Telomerase in Oncology
Cell Research (2002); 12(1):1-7 http://www.cell-research.com REVIEW Telomere and telomerase in oncology JIAO MU*, LI XIN WEI International Joint Cancer Institute, Second Military Medical University, Shanghai 200433, China ABSTRACT Shortening of the telomeric DNA at the chromosome ends is presumed to limit the lifespan of human cells and elicit a signal for the onset of cellular senescence. To continually proliferate across the senescent checkpoint, cells must restore and preserve telomere length. This can be achieved by telomerase, which has the reverse transcriptase activity. Telomerase activity is negative in human normal somatic cells but can be detected in most tumor cells. The enzyme is proposed to be an essential factor in cell immortalization and cancer progression. In this review we discuss the structure and function of telomere and telomerase and their roles in cell immortalization and oncogenesis. Simultaneously the experimental studies of telomerase assays for cancer detection and diagnosis are reviewed. Finally, we discuss the potential use of inhibitors of telomerase in anti-cancer therapy. Key words: Telomere, telomerase, cancer, telomerase assay, inhibitor. Telomere and cell replicative senescence base pairs of the end of telomeric DNA with each Telomeres, which are located at the end of round of DNA replication. Hence, the continual chromosome, are crucial to protect chromosome cycles of cell growth and division bring on progress- against degeneration, rearrangment and end to end ing telomere shortening[4]. Now it is clear that te- fusion[1]. Human telomeres are tandemly repeated lomere shortening is responsible for inducing the units of the hexanucleotide TTAGGG. The estimated senescent phenotype that results from repeated cell length of telomeric DNA varies from 2 to 20 kilo division, but the mechanism how a short telomere base pairs, depending on factors such as tissue type induces the senescence is still unknown. -

Ch 7 -Brock Information Flow in Biological Systems
Systems Microbiology Monday Oct 2 - Ch 7 -Brock Information flow in biological systems •• DNADNA replicationreplication •• TranscriptionTranscription •• TranslationTranslation Central Dogma DNA Replication Transcription Images removed due to copyright restrictions. RNA Reverse Transcription Translation Protein Flow of information replication DNA → DNA transcription ↓ RNA translation ↓ protein 5' end ring numbering system for -P-O-C deoxyribose 5’ -C O O 4’ 1’ P O- O 3’ 2’ C ssDNA 3’ end HO In a nucleotide, e.g., adenosine monophosphate (AMP), the base is bonded to a ribose sugar, which has a phosphate in ester linkage to the 5' hydroxyl. NH NH NH2 2 2 adenine N N N N N N N N N N N N H −2 HO O3P O CH CH 5' 2 O 2 O 4' H H 1' H H ribose H 3' 2' H H H OH OH OH OH adenine adenosine adenosine monophosphate (AMP) Nucleic acids have a NH2 backbone of adenine N N alternating Pi & ribose moieties. N NH − N 2 Phosphodiester 5' end O cytosine − 5' O P O CH N linkages form as the 2 O 4' 1' O H H ribose 5' phosphate of one N O H 3' 2' H nucleotide forms an O OH − 5' ester link with the 3' O P O CH 2 O OH of the adjacent O H H ribose nucleotide. H 3' H O OH − O P O (etc) nucleic acid 3' end O H N H O N Guanine Cytosine N H N N N N O H N Backbone Backbone Hydrogen H bond H O H N CH3 N Thymine N H N N Adenine N N Hydrogen O bond Backbone Backbone Figure by MIT OCW. -

Teacher Notes Science Nspired
DNA Replication TEACHER NOTES SCIENCE NSPIRED Science Objectives In this lesson, students will • Explore how the structure of DNA supports its semi-conservative replication • Identify the name and function of several key enzymes in DNA replication • Recognize the function of Okazaki fragments in DNA replication Vocabulary TI-Nspire™ Technology Skills: • Double helix • Leading strand • Download a TI-Nspire • Lagging strand • Polymerase I • Polymerase III • Helicase document • Base pairs • Okazaki Fragments • Open a document • Primase • Move between pages • Open Directions Box About the Lesson • Explore ‘Hot Spots’ • Using three simulations, students will interact with DNA replication to explore semi-conservative replication and identify Tech Tips: specific enzymes and their roles in replication. Assessments are Make sure that students know embedded in the activity to engage discussion and gauge how to move between pages by learning. pressing /¡ (left arrow) and • As a result, students will: /¢ (right arrow). • Learn the basic functions of the following DNA replication enzymes: helicase, primase, ligase, polymerase I and III. Lesson Materials: • Learn how the double helix reproduces in a semi- Student Activity • DNA_Replication_Student.do conservative fashion c • Learn the role and function of Okazaki fragments in • DNA_Replication_Student.pd replication of the lagging strand. f TI-Nspire document TI-Nspire™ Navigator™ • DNA_Replication.tns Not required. If using Navigator: • Send out the DNA_Replication.tns file. • Monitor student progress -

Dynamics of DNA Replication in a Eukaryotic Cell
Dynamics of DNA replication in a eukaryotic cell Thomas Kellya,1 and A. John Callegaria,2 aProgram in Molecular Biology, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY 10065 Contributed by Thomas Kelly, December 26, 2018 (sent for review October 30, 2018; reviewed by Paul Nurse and Nicholas Rhind) Each genomic locus in a eukaryotic cell has a distinct average time A deeper understanding of the dynamics of eukaryotic DNA of replication during S phase that depends on the spatial and replication will require a quantitative model describing the spatial temporal pattern of replication initiation events. Replication distribution of potential initiation sites and the time course of their timing can affect genomic integrity because late replication is activation during S phase. The fission yeast Schizosaccharomyces associated with an increased mutation rate. For most eukaryotes, pombe represents a useful system for developing such a model the features of the genome that specify the location and timing of because its genome has many characteristics in common with initiation events are unknown. To investigate these features for that of other eukaryotes. Early genetic studies identified seg- Schizosaccharomyces pombe the fission yeast, , we developed an ments of the S. pombe genome, called autonomously replicating integrative model to analyze large single-molecule and global ge- sequence (ars) elements, that function as origins of DNA repli- nomic datasets. The model provides an accurate description of the > S. pombe cation (14). These elements are large ( 1 kb) and rich in A and complex dynamics of DNA replication at high resolution. T residues but do not contain a common sequence motif (15, 16). -
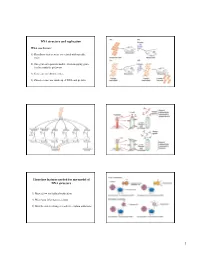
DNA Structure and Replication Three Key Features Needed for Any Model
DNA structure and replication What was known? 1) Hereditary factors were associated with specific traits 2) One-gene-one-protein model - from mapping genes for biosynthetic pathways 3) Genes are on chromosomes 4) Chromosomes are made up of DNA and protein Three key features needed for any model of DNA structure 1) Must allow for faithful replication 2) Must have information content 3) Must be able to change in order to explain mutations 1 Rosalind Franklin James Watson Francis Crick X-ray diffraction Maurice Wilkins Photograph of B-DNA Linus Pauling Nature 171, 737-738 (1953) © Macmillan Publishers Ltd. Molecular structure of Nucleic Acids WATSON, J. D. & CRICK, F. H. C. Features of the double helix A Structure for Deoxyribose Nucleic Acid 1) Two parallel strands We wish to suggest a structure for the salt of deoxyribose nucleic acid (D.N.A.). This structure has novel features which are of considerable biological interest………… 2) Bases held together by H-bonds 3) Phosphodiester backbone 4) The base is attached to the position 1 on the sugar 5) Base pair stack - provides Figure 1. This figure is purely diagrammatic. The two ribbons symbolize the two phophate-sugar chains, and the horizonal rods stability the pairs of bases holding the chains together. The vertical line marks the fibre axis. 6) Contains a major and …………….It has not escaped our notice that the specific pairing we minor groove have postulated immediately suggests a possible copying mechanism for the genetic material. Three key features needed for any model of DNA structure -

Processivity of DNA Polymerases: Two Mechanisms, One Goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2
Minireview 121 Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2 Replicative DNA polymerases are highly processive Processive DNA synthesis by cellular replicases and the enzymes that polymerize thousands of nucleotides without bacteriophage T4 replicase dissociating from the DNA template. The recently Until recently, the only mechanism for high processivity determined structure of the Escherichia coli bacteriophage that was understood in detail was that utilized by cellular T7 DNA polymerase suggests a unique mechanism that replicases and the replicase of bacteriophage T4. This underlies processivity, and this mechanism may generalize mechanism involves a ring-shaped protein called a ‘DNA to other replicative polymerases. sliding clamp’ that encircles the DNA and tethers the polymerase catalytic unit to the DNA [3,4]. The three- Addresses: 1Department of Molecular Biology, Memorial Sloan- dimensional structures of several sliding clamps have been Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, 2 determined: the eukaryotic proliferating cell nuclear USA and Laboratory of DNA Replication, Howard Hughes Medical β Institute, The Rockefeller University, 1230 York Avenue, New York, NY antigen (PCNA) [5,6]; the subunit of the prokaryotic 10021, USA. DNA polymerase III [7]; and the bacteriophage T4 gene 45 protein (gp45) (J Kuriyan, personal communication) *Corresponding author. (Figure 1). The overall structure of these clamps is very E-mail: [email protected] similar; the PCNA, β subunit and gp45 rings are super- Structure 15 February 1998, 6:121–125 imposable [8]. Each ring has similar dimensions and a http://biomednet.com/elecref/0969212600600121 central cavity large enough to accommodate duplex DNA (Figure 1). -

Primer Release Is the Rate-Limiting Event in Lagging-Strand Synthesis
Primer release is the rate-limiting event in lagging- strand synthesis mediated by the T7 replisome Alfredo J. Hernandeza, Seung-Joo Leea, and Charles C. Richardsona,1 aDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115 Contributed by Charles C. Richardson, April 18, 2016 (sent for review December 30, 2015; reviewed by Nicholas E. Dixon and I. Robert Lehman) DNA replication occurs semidiscontinuously due to the antiparallel but also for enabling the use of short oligoribonucleotides by T7 DNA strands and polarity of enzymatic DNA synthesis. Although DNA polymerase. Critically, the primase domain also fulfills two the leading strand is synthesized continuously, the lagging strand additional roles apart from primer synthesis: it prevents disso- is synthesized in small segments designated Okazaki fragments. ciation of the extremely short tetramer, stabilizing it with the Lagging-strand synthesis is a complex event requiring repeated template, and it secures it in the polymerase active site (10, 12). cycles of RNA primer synthesis, transfer to the lagging-strand Here we show that the rate-limiting step in initiation of Okazaki polymerase, and extension effected by cooperation between DNA fragments by the T7 replisome is primer release from the primase primase and the lagging-strand polymerase. We examined events domain of gp4. In the absence of gp2.5, an additional step, distinct controlling Okazaki fragment initiation using the bacteriophage from primer release, also limits primer extension. The presence of T7 replication system. Primer utilization by T7 DNA polymerase is gp2.5 promotes efficient primer formation and primer utilization. slower than primer formation.