Ambient Occlusion and Edge Cueing to Enhance Real Time Molecular Visualization
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Raytracing Prefiltered Occlusion for Aggregate Geometry
Raytracing Prefiltered Occlusion for Aggregate Geometry Dylan Lacewell1,2 Brent Burley1 Solomon Boulos3 Peter Shirley4,2 1 Walt Disney Animation Studios 2 University of Utah 3 Stanford University 4 NVIDIA Corporation Figure 1: Computing shadows using a prefiltered BVH is more efficient than using an ordinary BVH. (a) Using an ordinary BVH with 4 shadow rays per shading point requires 112 seconds for shadow rays, and produces significant visible noise. (b) Using a prefiltered BVH with 9 shadow rays requires 74 seconds, and visible noise is decreased. (c) Reducing noise to a similar level with an ordinary BVH requires 25 shadow rays and 704 seconds (about 9.5× slower). All images use 5 × 5 samples per pixel. The scene consists of about 2M triangles, each of which is semi-opaque (α = 0.85) to shadow rays. ABSTRACT aggregate geometry, it suffices to use a prefiltering algorithm that is We prefilter occlusion of aggregate geometry, e.g., foliage or hair, linear in the number of nodes in the BVH. Once built, a prefiltered storing local occlusion as a directional opacity in each node of a BVH does not need to be updated unless geometry changes. bounding volume hierarchy (BVH). During intersection, we termi- During rendering, we terminate shadow rays at some level of the nate rays early at BVH nodes based on ray differential, and compos- BVH, dependent on the differential [10] of each ray, and return the ite the stored opacities. This makes intersection cost independent of stored opacity at the node. The combined opacity of the ray is com- geometric complexity for rays with large differentials, and simulta- puted by compositing the opacities of one or more nodes that the neously reduces the variance of occlusion estimates. -

Fast Precomputed Ambient Occlusion for Proximity Shadows
INSTITUT NATIONAL DE RECHERCHE EN INFORMATIQUE ET EN AUTOMATIQUE Fast Precomputed Ambient Occlusion for Proximity Shadows Mattias Malmer — Fredrik Malmer — Ulf Assarsson — Nicolas Holzschuch N° 5779 Décembre 2005 Thème COG apport de recherche ISRN INRIA/RR--5779--FR+ENG ISSN 0249-6399 Fast Precomputed Ambient Occlusion for Proximity Shadows Mattias Malmer∗ , Fredrik Malmer∗ , Ulf Assarsson† ‡ , Nicolas Holzschuch‡ Thème COG — Systèmes cognitifs Projets ARTIS Rapport de recherche n° 5779 — Décembre 2005 — 19 pages Abstract: Ambient occlusion is used widely for improving the realism of real-time lighting simulations. We present a new, simple method for storing ambient occlusion values, that is very easy to implement and uses very little CPU and GPU resources. This method can be used to store and retrieve the percentage of occlusion, either alone or in combination with the average occluded direction. The former is cheaper in memory costs, while being slightly less accurate. The latter is slightly more expensive in memory, but gives more accurate results, especially when combining several occluders. The speed of our algorithm is independent of the complexity of either the occluder or the receiving scene. This makes the algorithm highly suitable for games and other real-time applications. Key-words: ∗ Syndicate Ent., Grevgatan 53, SE-114 58 Stockholm, Sweden. † Chalmers University of Technology, SE-412 96 Göteborg, Sweden. ‡ ARTIS/GRAVIR – IMAG INRIA Rhône-Alpes, France. Unité de recherche INRIA Rhône-Alpes 655, avenue de l’Europe, 38334 Montbonnot Saint Ismier (France) Téléphone : +33 4 76 61 52 00 — Télécopie +33 4 76 61 52 52 Fast Precomputed Ambient Occlusion for Proximity Shadows Résumé : L’Ambient Occlusion est fréquemment utilisée pour améliorer le réalisme de simulations de l’éclairage en temps-réel. -

A Web-Based 3D Molecular Structure Editor and Visualizer Platform
Mohebifar and Sajadi J Cheminform (2015) 7:56 DOI 10.1186/s13321-015-0101-7 SOFTWARE Open Access Chemozart: a web‑based 3D molecular structure editor and visualizer platform Mohamad Mohebifar* and Fatemehsadat Sajadi Abstract Background: Chemozart is a 3D Molecule editor and visualizer built on top of native web components. It offers an easy to access service, user-friendly graphical interface and modular design. It is a client centric web application which communicates with the server via a representational state transfer style web service. Both client-side and server-side application are written in JavaScript. A combination of JavaScript and HTML is used to draw three-dimen- sional structures of molecules. Results: With the help of WebGL, three-dimensional visualization tool is provided. Using CSS3 and HTML5, a user- friendly interface is composed. More than 30 packages are used to compose this application which adds enough flex- ibility to it to be extended. Molecule structures can be drawn on all types of platforms and is compatible with mobile devices. No installation is required in order to use this application and it can be accessed through the internet. This application can be extended on both server-side and client-side by implementing modules in JavaScript. Molecular compounds are drawn on the HTML5 Canvas element using WebGL context. Conclusions: Chemozart is a chemical platform which is powerful, flexible, and easy to access. It provides an online web-based tool used for chemical visualization along with result oriented optimization for cloud based API (applica- tion programming interface). JavaScript libraries which allow creation of web pages containing interactive three- dimensional molecular structures has also been made available. -

Combining Screen-Space Ambient Occlusion and Cartoon Rendering on Graphics Hardware
Combining Screen-Space Ambient Occlusion and Cartoon Rendering on Graphics Hardware Brett Lajzer Dan Nottingham Figure 1: Four visualizations of the same scene: a) no SSAO or outlining, b) SSAO, no outlines, c) no SSAO, outlines, d) SSAO and outlines 1. Motivation Screen-Space Ambient Occlusion Screen-space ambient occlusion (SSAO) is a further Methods for non-photorealistic rendering of 3D scenes have approximation of this technique, which was developed by become more popular in recent years for computer CryTek for their game Crysis and its engine. This version animation and games. We were interested in combining computes ambient occlusion for each pixel visible on the two particular NPR techniques: ambient occlusion and screen, by generating random points in the hemisphere cartoon shading. Ambient occlusion is an approach to around that pixel, and determining occlusion for each point global lighting that assumes that a point on the surface of by comparing its depth to a depth map of the scene. The an object receives less ambient light if there are many other sample is considered occluded if it is further from the objects occupying the space nearby in the hemisphere camera than the depth of the nearest visible object at that around that point. Screen-space ambient occlusion point, unless the difference in depth is greater than the approximates this on the GPU using the depth buffer to test sample radius. The advantage of this method is that it can occlusion of sample points. We combine this with cartoon be implemented on graphics hardware and run in real time, shading, which draws dark outlines on objects based on making it more suited to dynamic, interactive scenes due to depth and normal discontinuities, and thresholds lighting its dependence only upon screen resolution rather than intensity to several discreet values. -

Water and Salt at the Lipid-Solvent Interface
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School April 2019 Water and Salt at the Lipid-Solvent Interface James M. Kruczek University of South Florida, [email protected] Follow this and additional works at: https://scholarcommons.usf.edu/etd Part of the Physics Commons Scholar Commons Citation Kruczek, James M., "Water and Salt at the Lipid-Solvent Interface" (2019). Graduate Theses and Dissertations. https://scholarcommons.usf.edu/etd/8380 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Water and Salt at the Lipid-Solvent Interface by James M. Kruczek A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Applied Physics Department of Physics College of Arts and Sciences University of South Florida Major Professor: Sagar A. Pandit, Ph.D. Ullah, Ghanim, Ph.D. Robert S. Hoy, Ph.D. Jianjun Pan, Ph.D. Yicheng Tu, Ph.D. Date of Approval: March 26, 2019 Keywords: Lipid Bilayer, Ionic Solvents, Ether Lipids, Molecular Simulations Copyright ⃝c 2018, James M. Kruczek Dedication To my wife Nicole, without whom none of this would be possible. To my father Michael, who labored for his family till his last days. To my family, for all of their support. Acknowledgments The work presented in this document would not be possible without the assistance of many academic professionals. In particular, I would like to acknowledge my major advisor Dr. -

Fast Image-Based Ambient Occlusion IBAO
The International Journal of Virtual Reality, 2011, 10(4):61-65 61 Fast Image-Based Ambient Occlusion IBAO Robert Sajko and Zeljka Mihajlovic University of Zagreb, Faculty of Electrical Engineering and Computing,Department of Electronics, Microelectronics, Computer and Intelligent Systems Ambient lighting is an approximation of the light reflected from Abstract— The quality of computer rendering and perception other objects in the scene. Its presence reveals the spatial of realism greatly depend on the shading method used to relationship between objects, their shape, depth and surface implement the interaction of light with the surfaces of objects in a complexity details. Ambient lighting can be locally occluded scene. Ambient occlusion (AO) enhances the realistic impression by nearby object or a fold in the surface. Ambient occlusion of rendered objects and scenes. Properties that make Screen produces only subtle visual cues, however, they are very Space Ambient Occlusion (SSAO) interesting for real-time important in natural perception and thus also in a convincing, graphics are scene complexity independence, and support for fully dynamic scenes. However, there are also important issues with realistic lighting model. current approaches: poor texture cache use, introduction of noise, Modern consumer GPUs offer impressive computational and performance swings. power which has allowed the use of various techniques and In this paper, a straightforward solution is presented. Instead algorithms in real-time computer graphics that were previously of a traditional, geometry-based sampling method, a novel, possible in offline rendering only. One such technique is image-based sampling method is developed, coupled with a ambient occlusion, which approximates soft shadows due to revised heuristic function for computing occlusion. -

Neural Network Ambient Occlusion
Neural Network Ambient Occlusion Daniel Holden∗ Jun Saitoy Taku Komuraz University of Edinburgh Method Studios University of Edinburgh Figure 1: Comparison showing Neural Network Ambient Occlusion enabled and disabled inside a game engine. Abstract which is more accurate than existing techniques, has better perfor- mance, no user parameters other than the occlusion radius, and can We present Neural Network Ambient Occlusion (NNAO), a fast, ac- be computed in a single pass allowing it to be used as a drop-in curate screen space ambient occlusion algorithm that uses a neural replacement for existing techniques. network to learn an optimal approximation of the ambient occlu- sion effect. Our network is carefully designed such that it can be 2 Related Work computed in a single pass allowing it to be used as a drop-in re- placement for existing screen space ambient occlusion techniques. Screen Space Ambient Occlusion Screen Space Ambient Oc- clusion (SSAO) was first introduced by Mittring [2007] for use in Keywords: neural networks, machine learning, screen space am- Cryengine2. The approach samples around the depth buffer in a bient occlusion, SSAO, HBAO view space sphere and counts the number of points which are in- side the depth surface to estimate the occlusion. This method has seen wide adoption but often produces artifacts such as dark halos 1 Introduction around object silhouettes or white highlights on object edges. Fil- ion and McNaughton [2008] presented SSAO+, an extension which Ambient Occlusion is a key component in the lighting of a scene samples in a hemisphere oriented in the direction of the surface nor- but expensive to calculate. -

Tracking of Bacterial Metabolism with Azidated Precursors and Click- Chemistry”
Tracking of bacterial metabolism with azidated precursors and click-chemistry Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften vorgelegt beim Fachbereich für Biowissenschaften (15) der Johann Wolfang Goethe Universität in Frankfurt am Main von Alexander J. Pérez aus Nürnberg Frankfurt am Main 2015 Dekanin: Prof. Dr. Meike Piepenbring Gutachter: Prof. Dr. Helge B. Bode Zweitgutachter: Prof. Dr. Joachim W. Engels Datum der Disputation: 2 Danksagung Ich danke meinen Eltern für die stete und vielseitige Unterstützung, deren Umfang ich sehr zu schätzen weiß. Herrn Professor Dr. Helge B. Bode gilt mein besonderer Dank für die Übernahme als Doktorand und für die Gelegenheit meinen Horizont in diesen mich stets faszinierenden Themenbereich in dieser Tiefe erweitern zu lassen. Seine persönliche und fachliche Unterstützung bei der Projektwahl und der entsprechenden Umsetzung ist in dieser Form eine Seltenheit und ich bin mir dieser Tatsache voll bewusst. Gerade die zusätzlich erworbenen Kenntnisse im Bereich der Biologie, sowie der Wert interdisziplinärer Zusammenarbeit ist mir durch zahlreiche freundliche und wertvolle Mitglieder der Arbeitsgruppe bewusst geworden und viele zündenden Ideen wären ohne sie womöglich nie aufgekommen. Einen besonderen Dank möchte ich in diesem Kontext Wolfram Lorenzen und Sebastian Fuchs, die gerade in der Anfangszeit eine große Hilfe waren, ausdrücken. Dies gilt ebenso für die „N100-Crew“ und sämtliche Freunde, die in dieser Zeit zu mir standen und diesen Lebensabschnitt unvergesslich gemacht haben. -
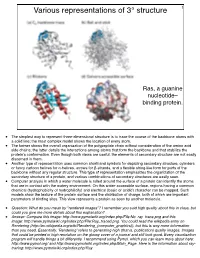
Various Representations of 3° Structure 1
Various representations of 3° structure 1 Ras, a guanine nucleotide– binding protein. • The simplest way to represent three-dimensional structure is to trace the course of the backbone atoms with a solid line; the most complex model shows the location of every atom. • The former shows the overall organization of the polypeptide chain without consideration of the amino acid side chains; the latter details the interactions among atoms that form the backbone and that stabilize the protein’s conformation. Even though both views are useful, the elements of secondary structure are not easily discerned in them. • Another type of representation uses common shorthand symbols for depicting secondary structure, cylinders or fancy cartoon helices for α-helices, arrows for β-strands, and a flexible string-like form for parts of the backbone without any regular structure. This type of representation emphasizes the organization of the secondary structure of a protein, and various combinations of secondary structures are easily seen. • Computer analysis in which a water molecule is rolled around the surface of a protein can identify the atoms that are in contact with the watery environment. On this water-accessible surface, regions having a common chemical (hydrophobicity or hydrophilicity) and electrical (basic or acidic) character can be mapped. Such models show the texture of the protein surface and the distribution of charge, both of which are important parameters of binding sites. This view represents a protein as seen by another molecule. • Question: What do you mean by "rendered images"? I remember you said high quality about this in class, but could you give me more details about this explanation? • Answer: Compare this image: http://www.pymolwiki.org/index.php/File:No_ray_trace.png and this image: http://www.pymolwiki.org/index.php/File:Ray_traced.png. -

Visualizing Protein Structures-Tools and Trends
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 12 January 2020 Visualizing protein structures - tools and trends 1,2 3 1,2 X. Martinez , M. Chavent , M. Baaden 1) CNRS, Université de Paris, UPR 9080, Laboratoire de Biochimie Théorique, 13 rue Pierre et Marie Curie, F-75005, Paris, France 2) Institut de Biologie Physico-Chimique-Fondation Edmond de Rotschild, PSL Research University, Paris, France 3) Institut de Pharmacologie et de Biologie Structurale IPBS, Université de Toulouse, CNRS, UPS, Toulouse, France Abstract Molecular visualisation is fundamental in the current scientific literature, textbooks and dissemination materials, forming an essential support for presenting results, reasoning on and formulating hypotheses related to molecular structure. Visual exploration has become easily accessible on a broad variety of platforms thanks to advanced software tools that render a great service to the scientific community. These tools are often developed across disciplines bridging computer science, biology and chemistry. Here we first describe a few Swiss Army knives geared towards protein visualisation for everyday use with an existing large user base, then focus on more specialised tools for peculiar needs that are not yet as broadly known. Our selection is by no means exhaustive, but reflects a diverse snapshot of scenarios that we consider informative for the reader. We end with an account of future trends and perspectives. Keywords Molecular Graphics, Protein visualization, Software tools, Virtual reality Introduction Many parts of science rely on a visualization-driven cycle of experimentation, reasoning, conjecture and validation, even more so in relation with structural biology and biophysics. Molecular visualization (1) in particular is now broadly used in many contexts, with the purpose of illustration in the scientific literature or the aim to gain insight about primary research data. -

Real-Time Ray Traced Ambient Occlusion and Animation Image Quality and Performance of Hardware- Accelerated Ray Traced Ambient Occlusion
DEGREE PROJECTIN COMPUTER SCIENCE AND ENGINEERING, SECOND CYCLE, 30 CREDITS STOCKHOLM, SWEDEN 2021 Real-time Ray Traced Ambient Occlusion and Animation Image quality and performance of hardware- accelerated ray traced ambient occlusion FABIAN WALDNER KTH ROYAL INSTITUTE OF TECHNOLOGY SCHOOL OF ELECTRICAL ENGINEERING AND COMPUTER SCIENCE Real-time Ray Traced Ambient Occlusion and Animation Image quality and performance of hardware-accelerated ray traced ambient occlusion FABIAN Waldner Master’s Programme, Industrial Engineering and Management, 120 credits Date: June 2, 2021 Supervisor: Christopher Peters Examiner: Tino Weinkauf School of Electrical Engineering and Computer Science Swedish title: Strålspårad ambient ocklusion i realtid med animationer Swedish subtitle: Bildkvalité och prestanda av hårdvaruaccelererad, strålspårad ambient ocklusion © 2021 Fabian Waldner Abstract | i Abstract Recently, new hardware capabilities in GPUs has opened the possibility of ray tracing in real-time at interactive framerates. These new capabilities can be used for a range of ray tracing techniques - the focus of this thesis is on ray traced ambient occlusion (RTAO). This thesis evaluates real-time ray RTAO by comparing it with ground- truth ambient occlusion (GTAO), a state-of-the-art screen space ambient occlusion (SSAO) method. A contribution by this thesis is that the evaluation is made in scenarios that includes animated objects, both rigid-body animations and skinning animations. This approach has some advantages: it can emphasise visual artefacts that arise due to objects moving and animating. Furthermore, it makes the performance tests better approximate real-world applications such as video games and interactive visualisations. This is particularly true for RTAO, which gets more expensive as the number of objects in a scene increases and have additional costs from managing the ray tracing acceleration structures. -

Engine for Molecule Visualization in a Web Browser
MASARYKOVA UNIVERZITA FAKULTA}w¡¢£¤¥¦§¨ INFORMATIKY !"#$%&'()+,-./012345<yA| Engine for Molecule Visualization in a Web Browser MASTER’S THESIS Jaromír Svoboda Brno, spring 2014 Declaration Hereby I declare, that this paper is my original authorial work, which I have worked out by my own. All sources, references and literature used or excerpted during elaboration of this work are properly cited and listed in complete reference to the due source. Jaromír Svoboda Advisor: RNDr. David Sehnal ii Acknowledgement I would like to thank my supervisor RNDr. David Sehnal for patient guidance and informed advice throughout writing this thesis. iii Abstract The main focus of this master’s thesis is the design and implementa- tion of lightweight molecular visualization engine (called LiveMol) in form of a JavaScript library. The engine utilizes widely adopted WebGL API to display GPU-accelerated graphics in web browsers. Due to the size of complex protein molecules, the primary goal is high performance. Furthermore, the design of LiveMol enables users to extend the core functionality by simply defining custom coloring schemes of molecule models or implement completely new visual- ization modes. iv Keywords molecular visualization, protein, secondary structure, WebGL, LiveMol v Contents I INTRODUCTION 1 1 Introduction ............................2 II CURRENT DEVELOPMENTS 3 2 Current Molecular Visualization Software ..........4 2.1 Desktop Applications ....................4 2.1.1 Visual Molecular Dynamic . .4 2.1.2 PyMOL . .4 2.1.3 RasMol/OpenRasMol . .5 2.1.4 BALL/BALLView . .5 2.1.5 Gabedit . .5 2.1.6 QuteMol . .5 2.1.7 Avogadro . .6 2.2 JavaScript-based Web Applications ............6 2.2.1 Jmol/JSmol .