Enzyme Nomenclature
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alternative Oxidase: a Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis During Abiotic and Biotic Stress in Plants
Int. J. Mol. Sci. 2013, 14, 6805-6847; doi:10.3390/ijms14046805 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Review Alternative Oxidase: A Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis during Abiotic and Biotic Stress in Plants Greg C. Vanlerberghe Department of Biological Sciences and Department of Cell and Systems Biology, University of Toronto Scarborough, 1265 Military Trail, Toronto, ON, M1C1A4, Canada; E-Mail: [email protected]; Tel.: +1-416-208-2742; Fax: +1-416-287-7676 Received: 16 February 2013; in revised form: 8 March 2013 / Accepted: 12 March 2013 / Published: 26 March 2013 Abstract: Alternative oxidase (AOX) is a non-energy conserving terminal oxidase in the plant mitochondrial electron transport chain. While respiratory carbon oxidation pathways, electron transport, and ATP turnover are tightly coupled processes, AOX provides a means to relax this coupling, thus providing a degree of metabolic homeostasis to carbon and energy metabolism. Beside their role in primary metabolism, plant mitochondria also act as “signaling organelles”, able to influence processes such as nuclear gene expression. AOX activity can control the level of potential mitochondrial signaling molecules such as superoxide, nitric oxide and important redox couples. In this way, AOX also provides a degree of signaling homeostasis to the organelle. Evidence suggests that AOX function in metabolic and signaling homeostasis is particularly important during stress. These include abiotic stresses such as low temperature, drought, and nutrient deficiency, as well as biotic stresses such as bacterial infection. This review provides an introduction to the genetic and biochemical control of AOX respiration, as well as providing generalized examples of how AOX activity can provide metabolic and signaling homeostasis. -

Articles Catalytic Cycling in Β-Phosphoglucomutase: a Kinetic
9404 Biochemistry 2005, 44, 9404-9416 Articles Catalytic Cycling in â-Phosphoglucomutase: A Kinetic and Structural Analysis†,‡ Guofeng Zhang, Jianying Dai, Liangbing Wang, and Debra Dunaway-Mariano* Department of Chemistry, UniVersity of New Mexico, Albuquerque, New Mexico 87131-0001 Lee W. Tremblay and Karen N. Allen* Department of Physiology and Biophysics, Boston UniVersity School of Medicine, Boston, Massachusetts 02118-2394 ReceiVed March 26, 2005; ReVised Manuscript ReceiVed May 18, 2005 ABSTRACT: Lactococcus lactis â-phosphoglucomutase (â-PGM) catalyzes the interconversion of â-D-glucose 1-phosphate (â-G1P) and â-D-glucose 6-phosphate (G6P), forming â-D-glucose 1,6-(bis)phosphate (â- G16P) as an intermediate. â-PGM conserves the core domain catalytic scaffold of the phosphatase branch of the HAD (haloalkanoic acid dehalogenase) enzyme superfamily, yet it has evolved to function as a mutase rather than as a phosphatase. This work was carried out to identify the structural basis underlying this diversification of function. In this paper, we examine â-PGM activation by the Mg2+ cofactor, â-PGM activation by Asp8 phosphorylation, and the role of cap domain closure in substrate discrimination. First, the 1.90 Å resolution X-ray crystal structure of the Mg2+-â-PGM complex is examined in the context of + + previously reported structures of the Mg2 -R-D-galactose-1-phosphate-â-PGM, Mg2 -phospho-â-PGM, and Mg2+-â-glucose-6-phosphate-1-phosphorane-â-PGM complexes to identify conformational changes that occur during catalytic turnover. The essential role of Asp8 in nucleophilic catalysis was confirmed by demonstrating that the D8A and D8E mutants are devoid of catalytic activity. -

Catalase and Oxidase Test
CATALASE TEST Catalase is the enzyme that breaks hydrogen peroxide (H 2O2) into H 2O and O 2. Hydrogen peroxide is often used as a topical disinfectant in wounds, and the bubbling that is seen is due to the evolution of O 2 gas. H 2O2 is a potent oxidizing agent that can wreak havoc in a cell; because of this, any cell that uses O 2 or can live in the presence of O 2 must have a way to get rid of the peroxide. One of those ways is to make catalase. PROCEDURE a. Place a small amount of growth from your culture onto a clean microscope slide. If using colonies from a blood agar plate, be very careful not to scrape up any of the blood agar— blood cells are catalase positive and any contaminating agar could give a false positive. b. Add a few drops of H 2O2 onto the smear. If needed, mix with a toothpick. DO NOT use a metal loop or needle with H 2O2; it will give a false positive and degrade the metal. c. A positive result is the rapid evolution of O 2 as evidenced by bubbling. d. A negative result is no bubbles or only a few scattered bubbles. e. Dispose of your slide in the biohazard glass disposal container. Dispose of any toothpicks in the Pipet Keeper. OXIDASE TEST Basically, this is a test to see if an organism is an aerobe. It is a check for the presence of the electron transport chain that is the final phase of aerobic respiration. -

Glucose Oxidase
Catalog Number: 100289, 100330, 195196 Glucose Oxidase Molecular Weight: ~160,0001 CAS #: 9001-34-0 Physical Description: Yellow lyophilized powder Source: Aspergillus niger Isoelectric Point: 4.2 Michaelis Constants: phosphate buffer, pH 5.6; 25°C, air: Glucose: 3.3 x 10-2 mol/l(2) Glucose: 1.1 x 10-1 mol/l(3) 2-Deoxyglucose: 2.5 x 10-2 mol/l O2: 2.0 x 10-4 mol/l Structure: The enzyme contains 2 moles FAD/mole GOD. Inhibitors: Ag+, Hg2+, Cu2+ (4), 4-chloromercuribenzoate, D-arabinose (50%). FAD binding is inhibited by several nucleotides.5 pH Optimum: 6.5 pH Stability: 8.0 pH Range: 4-7 Thermal Stability: Below 40°C Description: Glucose oxidase is an FAD-containing glycoprotein. The enzyme is specific for b-D-glucose. O2 can be replaced by hydrogen acceptors such as 2.6-dichlorophenol indophenol. Relative Rates: D-glucose, 100; D-mannose, 20; 2-Deoxy-D-glucose, 20; negligible on other hexoses. Solubility: Dissolves readily at 5 mg/ml in 0.1 M potassium phosphate pH 7.0, giving a clear, yellow solution, also soluble in water Assay Procedure 1: Unit Definition: One unit of glucose oxidase is the activity which causes the liberation of 1 micromole of H2O2 per minute at 25°C and pH 7.0 under the specified conditions. Reaction: Reagents: 1. 0.1 M Phosphate Buffer, pH 6.8: dissolve 6.8 g of potassium phosphate, monobasic, anhydrous and 7.1 g of sodium phosphate, dibasic, anhydrous in about 800 ml of deionized water. Adjust the pH to 6.8 ± 0.05 @ 25°C with 1 N HCl or 1N NaOH if necessary. -

Isocitrate Dehydrogenase Activity Assay Kit (MAK062)
Isocitrate Dehydrogenase Activity Assay Kit Catalog Number MAK062 Storage Temperature –20 C TECHNICAL BULLETIN Product Description Developer 1 vl Isocitrate dehydrogenase (IDH) catalyzes the Catalog Number MAK062E conversion of isocitrate to -ketoglutarate. In eukaryotes, there are three isozymes of IDH, the IDH Positive Control (NADP+) 20 L mitochondrial IDH2 and IDH3, and the cytoplasmic/ Catalog Number MAK062F peroxisomal IDH1. All three IDH family members require the presence of a divalent cation (Mg2+ or Mn2+) NADH Standard, 0.5 mole 1 vl and either the electron-accepting cofactor NADP+ (IDH1 Catalog Number MAK062G and IDH2) or NAD+ (IDH3) for their enzymatic activity. IDH1 and IDH2 mutations resulting in neomorphic Reagents and Equipment Required but Not enzymatic activity are found in certain cancers such as Provided. glioblastoma, acute myeloid leukemia, and colon 96 well flat-bottom plate – It is recommended to use cancer. This neoactivity shows a change in the clear plates for colorimetric assays. substrate specificity resulting in the conversion of Spectrophotometric multiwell plate reader -ketoglutarate to 2-hydroxyglutarate. Mutations in IDH family members are also associated with Ollier disease Precautions and Disclaimer and Maffucci syndrome. This product is for R&D use only, not for drug, household, or other uses. Please consult the Material The Isocitrate Dehydrogenase Activity Assay kit Safety Data Sheet for information regarding hazards provides a simple and direct procedure for measuring and safe handling practices. + + + NADP -dependent, NAD -dependent, or both NADP + and NAD -dependent IDH activity in a variety of Preparation Instructions samples. IDH activity is determined using isocitrate as Briefly centrifuge vials before opening. -

Isocitrate Dehydrogenase 1 (NADP+) (I5036)
Isocitrate Dehydrogenase 1 (NADP+), human recombinant, expressed in Escherichia coli Catalog Number I5036 Storage Temperature –20 °C CAS RN 9028-48-2 IDH1 and IDH2 have frequent genetic alterations in EC 1.1.1.42 acute myeloid leukemia4 and better understanding of Systematic name: Isocitrate:NADP+ oxidoreductase these mutations may lead to an improvement of (decarboxylating) individual cancer risk assessment.6 In addition other studies have shown loss of IDH1 in bladder cancer Synonyms: IDH1, cytosolic NADP(+)-dependent patients during tumor development suggesting this may isocitrate dehydrogenase, isocitrate:NADP+ be involved in tumor progression and metastasis.7 oxidoreductase (decarboxylating), Isocitric Dehydrogenase, ICD1, PICD, IDPC, ICDC, This product is lyophilized from a solution containing oxalosuccinate decarboxylase Tris-HCl, pH 8.0, with trehalose, ammonium sulfate, and DTT. Product Description Isocitrate dehydrogenase (NADP+) [EC 1.1.1.42] is a Purity: ³90% (SDS-PAGE) Krebs cycle enzyme, which converts isocitrate to a-ketoglutarate. The flow of isocitrate through the Specific activity: ³80 units/mg protein glyoxylate bypass is regulated by phosphorylation of isocitrate dehydrogenase, which competes for a Unit definition: 1 unit corresponds to the amount of 1 common substrate (isocitrate) with isocitrate lyase. enzyme, which converts 1 mmole of DL-isocitrate to The activity of the enzyme is dependent on the a-ketoglutarate per minute at pH 7.4 and 37 °C (NADP formation of a magnesium or manganese-isocitrate as cofactor). The activity is measured by observing the 2 complex. reduction of NADP to NADPH at 340 nm in the 7 presence of 4 mM DL-isocitrate and 2 mM MnSO4. -

Genetic Variants of Microsomal Epoxide Hydrolase and Glutamate-Cysteine Ligase In
ERJ Express. Published on July 9, 2008 as doi: 10.1183/09031936.00065308 Genetic variants of microsomal epoxide hydrolase and glutamate-cysteine ligase in COPD Running Title: EPHX1 and GCL variation in COPD Sally Chappell1, Leslie Daly2, Kevin Morgan1, Tamar Guetta-Baranes1, Josep Roca3, Roberto Rabinovich3, Juzer Lotya2,Ann B. Millar4, Seamas C. Donnelly5, Vera Keatings6, William MacNee7, Jan Stolk8, Pieter S. Hiemstra8, Massimo Miniati9, Simonetta Monti9 Clare M. O’Connor5,10 and Noor Kalsheker1,10. 1 The University of Nottingham. Division of Clinical Chemistry, Molecular Medical Sciences, Institute of Genetics, University Hospital, Queens Medical Centre, Nottingham, NG7 2UH, UK 2 University College Dublin. UCD School of Public Health & Population Science, UCD, Belfield, Dublin 4, IRELAND 3 Hospital Clinico y Provincial de Barcelona. Service de Pneumologia, Hospital Clinic, Villarroel, 170, 08036, Barcelona, SPAIN. 4 University of Bristol. Lung Research Group, Department of Clinical Science at North Bristol, Southmead Hospital, Westbury on Trym, Bristol BS10 5NB, UK 5 University College Dublin. UCD School of Medicine and Medical Science, The Conway Institute, UCD, Belfield, Dublin 4, IRELAND 6 Letterkenny General Hospital, Letterkenny, Co. Donegal, Ireland 1 Copyright 2008 by the European Respiratory Society. 7 ELEGI Colt Laboratories, MRC Centre for Inflammation Research, Level 2, Room C2.29, The Queen’s Medical Research Institute, 47 Little France Crescent, Edinburgh EH16 4TJ. 8 Leiden University Medical Center, Department of Pulmonology (C3-P), Albinusdreef 2, P.O. Box 9600, 2300 RC Leiden, THE NETHERLANDS 9 CNR Institute of Clinical Physiology, Via G. Moruzzi 1-56124, Pisa, ITALY 10 Joint senior authors. Corresponding author: Professor Noor Kalsheker, Division of Clinical Chemistry, University Hospital, Nottingham, NG7 2UH, UK. -
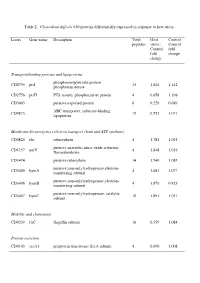
Table 2: Clostridium Difficile 630 Proteins Differentially Expressed in Response to Heat Stress
Table 2: Clostridium difficile 630 proteins differentially expressed in response to heat stress. Locus Gene name Description Total Heat Control : peptides stress : Control Control fold fold change change Transport/binding proteins and lipoproteins phosphoenolpyruvate-protein CD2755 ptsI 15 1.644 1.142 phosphotransferase CD2756 ptsH PTS system, phosphocarrier protein 4 0.658 1.198 CD3043 putative exported protein 6 0.229 0.689 ABC transporter, substrate-binding CD0873 15 0.521 1.071 lipoprotein Membrane bioenergetics (electron transport chain and ATP synthase) CD0825 rbr rubrerythrin 4 1.781 1.035 putative anaerobic nitric oxide reductase CD1157 norV 4 1.848 1.016 flavorubredoxin CD1474 putative ruberythrin 14 1.540 1.085 putative iron-only hydrogenase,electron- CD3405 hymA 4 1.681 1.077 transferring subunit putative iron-only hydrogenase,electron- CD3406 hymB 4 1.870 0.923 transferring subunit putative iron-only hydrogenase, catalytic CD3407 hymC 10 1.891 1.051 subunit Mobility and chemotaxis CD0239 fliC flagellin subunit 16 0.559 1.084 Protein secretion CD0143 secA1 preprotein translocase SecA subunit 4 0.690 1.008 Specific pathways putative oxidoreductase, thiamine diP-binding CD0116 13 1.628 1.121 subunit putative oxidoreductase, acetyl-CoA synthase CD0174 4 2.687 1.055 subunit CD0790 putative NUDIX-family hydrolase 8 1.491 1.32 inosine-uridine preferring nucleoside CD1682 iunH 4 0.518 0.984 hydrolase CD2181 putative aromatic compounds hydrolase 7 0372 0.909 CD2342 sucD succinate-semialdehyde dehydrogenase 6 0.651 0.906 Metabolism -

(LCHAD) Deficiency / Mitochondrial Trifunctional Protein (MTF) Deficiency
Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / Mitochondrial trifunctional protein (MTF) deficiency Contact details Introduction Regional Genetics Service Long chain acyl-CoA dehydrogenase (LCHAD) deficiency / mitochondrial trifunctional Levels 4-6, Barclay House protein (MTF) deficiency is an autosomal recessive disorder of mitochondrial beta- 37 Queen Square oxidation of fatty acids. The mitochondrial trifunctional protein is composed of 4 alpha London, WC1N 3BH and 4 beta subunits, which are encoded by the HADHA and HADHB genes, respectively. It is characterized by early-onset cardiomyopathy, hypoglycemia, T +44 (0) 20 7762 6888 neuropathy, and pigmentary retinopathy, and sudden death. There is also an infantile F +44 (0) 20 7813 8578 onset form with a hepatic Reye-like syndrome, and a late-adolescent onset form with primarily a skeletal myopathy. Tandem mass spectrometry of organic acids in urine, Samples required and carnitines in blood spots, allows the diagnosis to be unequivocally determined. An 5ml venous blood in plastic EDTA additional clinical complication can occur in the pregnant mothers of affected fetuses; bottles (>1ml from neonates) they may experience maternal acute fatty liver of pregnancy (AFLP) syndrome or Prenatal testing must be arranged hypertension/haemolysis, elevated liver enzymes and low platelets (HELLP) in advance, through a Clinical syndrome. Genetics department if possible. The genes encoding the HADHA and HADHB subunits are located on chromosome Amniotic fluid or CV samples 2p23.3. The pathogenic -

Enzyme Phosphatidylserine Synthase (Saccharomyces Cerevisae/Chol Gene/Transformation) V
Proc. Nati. Acad. Sci. USA Vol. 80, pp. 7279-7283, December 1983 Genetics Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase (Saccharomyces cerevisae/CHOl gene/transformation) V. A. LETTS*, L. S. KLIG*, M. BAE-LEEt, G. M. CARMANt, AND S. A. HENRY* *Departments of Genetics and Molecular Biology, Albert Einstein College of Medicine, Bronx, NY 10461; and tDepartment of Food Science, Cook College, New Jersey Agricultural Experimental Station, Rutgers University, New Brunswick, NJ 08903 Communicated by Frank Lilly, August 11, 1983 ABSTRACT The structural gene (CHOI) for phosphatidyl- Mammals, for example, synthesize PtdSer by an exchange re- serine synthase (CDPdiacylglycerol:L-serine O-phosphatidyl- action with PtdEtn (9). However, PtdSer synthase is found in transferase, EC 2.7.8.8) was isolated by genetic complementation E. coli and indeed the structural gene for the E. coli enzyme has in Saccharomyces cerevmae from a bank of yeast genomic DNA been cloned (10). Thus, cloning of the structural gene for the on a chimeric plasmid. The cloned DNA (4.0 kilobases long) was yeast enzyme will permit a detailed comparison of the structure shown to represent a unique sequence in the yeast genome. The and function of prokaryotic and eukaryotic genes and gene DNA sequence on an integrative plasmid was shown to recombine products. The availability of a clone of the CHOI gene will per- into the CHOi locus, confwrming its genetic identity. The chol yeast mit analysis of its regulation at the transcriptional level. Fur- strain transformed with this gene on an autonomously replicating thermore, the cloning of the CHOI gene provides us with the plasmid had significantly increased activity of the regulated mem- the levels of PtdSer synthase in the cell, brane-associated enzyme phosphatidylserine synthase. -

Fructose As an Endogenous Toxin
HEPATOCYTE MOLECULAR CYTOTOXIC MECHANISM STUDY OF FRUCTOSE AND ITS METABOLITES INVOLVED IN NONALCOHOLIC STEATOHEPATITIS AND HYPEROXALURIA By Yan (Cynthia) Feng A thesis submitted in the conformity with the requirements for the degree of Master of Science Graduate Department of Pharmaceutical Sciences University of Toronto © Copyright by Yan (Cynthia) Feng 2010 ABSTRACT HEPATOCYTE MOLECULAR CYTOTOXIC MECHANISM STUDY OF FRUCTOSE AND ITS METABOLITES INVOLVED IN NONALCOHOLIC STEATOHEPATITIS AND HYPEROXALURIA Yan (Cynthia) Feng Master of Science, 2010 Department of Pharmaceutical Sciences University of Toronto High chronic fructose consumption is linked to a nonalcoholic steatohepatitis (NASH) type of hepatotoxicity. Oxalate is the major endpoint of fructose metabolism, which accumulates in the kidney causing renal stone disease. Both diseases are life-threatening if not treated. Our objective was to study the molecular cytotoxicity mechanisms of fructose and some of its metabolites in the liver. Fructose metabolites were incubated with primary rat hepatocytes, but cytotoxicity only occurred if the hepatocytes were exposed to non-toxic amounts of hydrogen peroxide such as those released by activated immune cells. Glyoxal was most likely the endogenous toxin responsible for fructose induced toxicity formed via autoxidation of the fructose metabolite glycolaldehyde catalyzed by superoxide radicals, or oxidation by Fenton’s hydroxyl radicals. As for hyperoxaluria, glyoxylate was more cytotoxic than oxalate presumably because of the formation of condensation product oxalomalate causing mitochondrial toxicity and oxidative stress. Oxalate toxicity likely involved pro-oxidant iron complex formation. ii ACKNOWLEDGEMENTS I would like to dedicate this thesis to my family. To my parents, thank you for the sacrifices you have made for me, thank you for always being there, loving me and supporting me throughout my life. -

Aminoacyl-Trna Synthetases: Versatile Players in the Changing Theater of Translation
Downloaded from rnajournal.cshlp.org on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press RNA (2002), 8:1363–1372+ Cambridge University Press+ Printed in the USA+ Copyright © 2002 RNA Society+ DOI: 10+1017/S1355838202021180 MEETING REVIEW Aminoacyl-tRNA synthetases: Versatile players in the changing theater of translation CHRISTOPHER FRANCKLYN,1 JOHN J. PERONA,2 JOERN PUETZ,3 and YA-MING HOU4 1 Department of Biochemistry, University of Vermont, Burlington, Vermont 05405, USA 2 Department of Chemistry and Biochemistry, University of California, Santa Barbara, California 93106-9510, USA 3 UPR9002 du Centre National de la Recherche Scientifique, Institut de Biologie Moléculaire et Cellulaire, Strasbourg, 67084 Cedex, France 4 Department of Biochemistry, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, USA ABSTRACT Aminoacyl-tRNA synthetases attach amino acids to the 39 termini of cognate tRNAs to establish the specificity of protein synthesis. A recent Asilomar conference (California, January 13–18, 2002) discussed new research into the structure–function relationship of these crucial enzymes, as well as a multitude of novel functions, including par- ticipation in amino acid biosynthesis, cell cycle control, RNA splicing, and export of tRNAs from nucleus to cytoplasm in eukaryotic cells. Together with the discovery of their role in the cellular synthesis of proteins to incorporate selenocysteine and pyrrolysine, these diverse functions of aminoacyl-tRNA synthetases underscore the flexibility and adaptability