Sequencing the Fungal Tree of Life
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
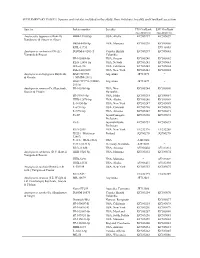
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Genome Sequence Analysis of Auricularia Heimuer Combined with Genetic Linkage Map
Journal of Fungi Article Genome Sequence Analysis of Auricularia heimuer Combined with Genetic Linkage Map Ming Fang 1, Xiaoe Wang 2, Ying Chen 2, Peng Wang 2, Lixin Lu 2, Jia Lu 2, Fangjie Yao 1,2,* and Youmin Zhang 1,* 1 Lab of genetic breeding of edible mushromm, Horticultural, College of Horticulture, Jilin Agricultural University, Changchun 130118, China; [email protected] 2 Engineering Research Centre of Chinese Ministry of Education for Edible and Medicinal Fungi, Jilin Agricultural University, Changchun 130118, China; [email protected] (X.W.); [email protected] (Y.C.); [email protected] (P.W.); [email protected] (L.L.); [email protected] (J.L.) * Correspondence: [email protected] (F.Y.); [email protected] (Y.Z.) Received: 3 March 2020; Accepted: 12 March 2020; Published: 16 March 2020 Abstract: Auricularia heimuer is one of the most popular edible fungi in China. In this study, the whole genome of A. heimuer was sequenced on the Illumina HiSeq X system and compared with other mushrooms genomes. As a wood-rotting fungus, a total of 509 carbohydrate-active enzymes (CAZymes) were annotated in order to explore its potential capabilities on wood degradation. The glycoside hydrolases (GH) family genes in the A. heimuer genome were more abundant than the genes in the other 11 mushrooms genomes. The A. heimuer genome contained 102 genes encoding class III, IV, and V ethanol dehydrogenases. Evolutionary analysis based on 562 orthologous single-copy genes from 15 mushrooms showed that Auricularia formed an early independent branch of Agaricomycetes. The mating-type locus of A. heimuer was located on linkage group 8 by genetic linkage analysis. -

Studies on Ear Fungus-Auricularia from the Woodland of Nameri National Park, Sonitpur District, Assam
International Journal of Interdisciplinary and Multidisciplinary Studies (IJIMS), 2014, Vol 1, No.5, 262-265. 262 Available online at http://www.ijims.com ISSN: 2348 – 0343 Studies on Ear Fungus-Auricularia from the Woodland of Nameri National Park, Sonitpur District, Assam. M.P. Choudhury1*, Dr.T.C Sarma2 1.Department of Botany, Nowgong College, Nagaon -782001, Assam, India. 2.Department of Botany, Gauhati University,Guwahati-7810 14, Assam, India. *Corresponding author: M.P. Choudhury Abstract Auricularia is the genus of the order Auriculariales with more than 10 species. It is also called ear fungus due to its morphological similarities with human ear and has considerable mythological importance. Auricularia auricula is the type species of the order Auriculariales. Different species of Auricularia are edible and some have medicinal importance and still investigations are going on other species to find out their medicinal properties. Extensive woodland of Nameri National Park provides ideal condition for the growth of different species of Auricularia. In this context the present study has been undertaken to study the taxonomy and diversity of different species of Auricularia and bring together information of its ethenomycological uses. As a result of field and laboratory study four different species of Auricularia were collected of which 3 species were identified and one species remain unidentified. Key Words: Auricularia, Taxonomy, Diversity, Nameri National Park. Introduction Auricularia belongs to the order Auriculariales is the largest genus of jelly fungi. They are among the most common and widely distributed members of macrofungi, which generally occurs as saprophytes on wood, logs, branch and twigs causing severe degrees of white rotting of forest trees. -

Annotated Check List and Host Index Arizona Wood
Annotated Check List and Host Index for Arizona Wood-Rotting Fungi Item Type text; Book Authors Gilbertson, R. L.; Martin, K. J.; Lindsey, J. P. Publisher College of Agriculture, University of Arizona (Tucson, AZ) Rights Copyright © Arizona Board of Regents. The University of Arizona. Download date 28/09/2021 02:18:59 Link to Item http://hdl.handle.net/10150/602154 Annotated Check List and Host Index for Arizona Wood - Rotting Fungi Technical Bulletin 209 Agricultural Experiment Station The University of Arizona Tucson AÏfJ\fOTA TED CHECK LI5T aid HOST INDEX ford ARIZONA WOOD- ROTTlNg FUNGI /. L. GILßERTSON K.T IyIARTiN Z J. P, LINDSEY3 PRDFE550I of PLANT PATHOLOgY 2GRADUATE ASSISTANT in I?ESEARCI-4 36FZADAATE A5 S /STANT'" TEACHING Z z l'9 FR5 1974- INTRODUCTION flora similar to that of the Gulf Coast and the southeastern United States is found. Here the major tree species include hardwoods such as Arizona is characterized by a wide variety of Arizona sycamore, Arizona black walnut, oaks, ecological zones from Sonoran Desert to alpine velvet ash, Fremont cottonwood, willows, and tundra. This environmental diversity has resulted mesquite. Some conifers, including Chihuahua pine, in a rich flora of woody plants in the state. De- Apache pine, pinyons, junipers, and Arizona cypress tailed accounts of the vegetation of Arizona have also occur in association with these hardwoods. appeared in a number of publications, including Arizona fungi typical of the southeastern flora those of Benson and Darrow (1954), Nichol (1952), include Fomitopsis ulmaria, Donkia pulcherrima, Kearney and Peebles (1969), Shreve and Wiggins Tyromyces palustris, Lopharia crassa, Inonotus (1964), Lowe (1972), and Hastings et al. -

Molecular Evolution and Functional Divergence of Tubulin Superfamily In
OPEN Molecular evolution and functional SUBJECT AREAS: divergence of tubulin superfamily in the FUNGAL GENOMICS MOLECULAR EVOLUTION fungal tree of life FUNGAL BIOLOGY Zhongtao Zhao1*, Huiquan Liu1*, Yongping Luo1, Shanyue Zhou2, Lin An1, Chenfang Wang1, Qiaojun Jin1, Mingguo Zhou3 & Jin-Rong Xu1,2 Received 18 July 2014 1 NWAFU-PU Joint Research Center, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Plant Protection, 2 Accepted Northwest A&F University, Yangling, Shaanxi 712100, China, Department of Botany and Plant Pathology, Purdue University, West 3 22 September 2014 Lafayette, IN 47907, USA, College of Plant Protection, Nanjing Agricultural University, Key Laboratory of Integrated Management of Crop Diseases and Pests, Ministry of Education, Key Laboratory of Pesticide, Nanjing, Jiangsu 210095, China. Published 23 October 2014 Microtubules are essential for various cellular activities and b-tubulins are the target of benzimidazole fungicides. However, the evolution and molecular mechanisms driving functional diversification in fungal tubulins are not clear. In this study, we systematically identified tubulin genes from 59 representative fungi Correspondence and across the fungal kingdom. Phylogenetic analysis showed that a-/b-tubulin genes underwent multiple requests for materials independent duplications and losses in different fungal lineages and formed distinct paralogous/ should be addressed to orthologous clades. The last common ancestor of basidiomycetes and ascomycetes likely possessed two a a a b b b a J.-R.X. (jinrong@ paralogs of -tubulin ( 1/ 2) and -tubulin ( 1/ 2) genes but 2-tubulin genes were lost in basidiomycetes and b2-tubulin genes were lost in most ascomycetes. Molecular evolutionary analysis indicated that a1, a2, purdue.edu) and b2-tubulins have been under strong divergent selection and adaptive positive selection. -

Agaricales, Basidiomycota) Occurring in Punjab, India
Current Research in Environmental & Applied Mycology 5 (3): 213–247(2015) ISSN 2229-2225 www.creamjournal.org Article CREAM Copyright © 2015 Online Edition Doi 10.5943/cream/5/3/6 Ecology, Distribution Perspective, Economic Utility and Conservation of Coprophilous Agarics (Agaricales, Basidiomycota) Occurring in Punjab, India Amandeep K1*, Atri NS2 and Munruchi K2 1Desh Bhagat College of Education, Bardwal–Dhuri–148024, Punjab, India. 2Department of Botany, Punjabi University, Patiala–147002, Punjab, India. Amandeep K, Atri NS, Munruchi K 2015 – Ecology, Distribution Perspective, Economic Utility and Conservation of Coprophilous Agarics (Agaricales, Basidiomycota) Occurring in Punjab, India. Current Research in Environmental & Applied Mycology 5(3), 213–247, Doi 10.5943/cream/5/3/6 Abstract This paper includes the results of eco-taxonomic studies of coprophilous mushrooms in Punjab, India. The information is based on the survey to dung localities of the state during the various years from 2007-2011. A total number of 172 collections have been observed, growing as saprobes on dung of various domesticated and wild herbivorous animals in pastures, open areas, zoological parks, and on dung heaps along roadsides or along village ponds, etc. High coprophilous mushrooms’ diversity has been established and a number of rare and sensitive species recorded with the present study. The observed collections belong to 95 species spread over 20 genera and 07 families of the order Agaricales. The present paper discusses the distribution of these mushrooms in Punjab among different seasons, regions, habitats, and growing habits along with their economic utility, habitat management and conservation. This is the first attempt in which various dung localities of the state has been explored systematically to ascertain the diversity, seasonal availability, distribution and ecology of coprophilous mushrooms. -

Major Clades of Agaricales: a Multilocus Phylogenetic Overview
Mycologia, 98(6), 2006, pp. 982–995. # 2006 by The Mycological Society of America, Lawrence, KS 66044-8897 Major clades of Agaricales: a multilocus phylogenetic overview P. Brandon Matheny1 Duur K. Aanen Judd M. Curtis Laboratory of Genetics, Arboretumlaan 4, 6703 BD, Biology Department, Clark University, 950 Main Street, Wageningen, The Netherlands Worcester, Massachusetts, 01610 Matthew DeNitis Vale´rie Hofstetter 127 Harrington Way, Worcester, Massachusetts 01604 Department of Biology, Box 90338, Duke University, Durham, North Carolina 27708 Graciela M. Daniele Instituto Multidisciplinario de Biologı´a Vegetal, M. Catherine Aime CONICET-Universidad Nacional de Co´rdoba, Casilla USDA-ARS, Systematic Botany and Mycology de Correo 495, 5000 Co´rdoba, Argentina Laboratory, Room 304, Building 011A, 10300 Baltimore Avenue, Beltsville, Maryland 20705-2350 Dennis E. Desjardin Department of Biology, San Francisco State University, Jean-Marc Moncalvo San Francisco, California 94132 Centre for Biodiversity and Conservation Biology, Royal Ontario Museum and Department of Botany, University Bradley R. Kropp of Toronto, Toronto, Ontario, M5S 2C6 Canada Department of Biology, Utah State University, Logan, Utah 84322 Zai-Wei Ge Zhu-Liang Yang Lorelei L. Norvell Kunming Institute of Botany, Chinese Academy of Pacific Northwest Mycology Service, 6720 NW Skyline Sciences, Kunming 650204, P.R. China Boulevard, Portland, Oregon 97229-1309 Jason C. Slot Andrew Parker Biology Department, Clark University, 950 Main Street, 127 Raven Way, Metaline Falls, Washington 99153- Worcester, Massachusetts, 01609 9720 Joseph F. Ammirati Else C. Vellinga University of Washington, Biology Department, Box Department of Plant and Microbial Biology, 111 355325, Seattle, Washington 98195 Koshland Hall, University of California, Berkeley, California 94720-3102 Timothy J. -

GFS Fungal Remains from Late Neogene Deposits at the Gray
GFS Mycosphere 9(5): 1014–1024 (2018) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/9/5/5 Fungal remains from late Neogene deposits at the Gray Fossil Site, Tennessee, USA Worobiec G1, Worobiec E1 and Liu YC2 1 W. Szafer Institute of Botany, Polish Academy of Sciences, Lubicz 46, PL-31-512 Kraków, Poland 2 Department of Biological Sciences and Office of Research & Sponsored Projects, California State University, Fullerton, CA 92831, U.S.A. Worobiec G, Worobiec E, Liu YC 2018 – Fungal remains from late Neogene deposits at the Gray Fossil Site, Tennessee, USA. Mycosphere 9(5), 1014–1024, Doi 10.5943/mycosphere/9/5/5 Abstract Interesting fungal remains were encountered during palynological investigation of the Neogene deposits at the Gray Fossil Site, Washington County, Tennessee, USA. Both Cephalothecoidomyces neogenicus and Trichothyrites cf. padappakarensis are new for the Neogene of North America, while remains of cephalothecoid fungus Cephalothecoidomyces neogenicus G. Worobiec, Neumann & E. Worobiec, fragments of mantle tissue of mycorrhizal Cenococcum and sporocarp of epiphyllous Trichothyrites cf. padappakarensis (Jain & Gupta) Kalgutkar & Jansonius were reported. Remains of mantle tissue of Cenococcum for the fossil state are reported for the first time. The presence of Cephalothecoidomyces, Trichothyrites, and other fungal remains previously reported from the Gray Fossil Site suggest warm and humid palaeoclimatic conditions in the southeast USA during the late Neogene, which is in accordance with data previously obtained from other palaeontological analyses at the Gray Fossil Site. Key words – Cephalothecoid fungus – Epiphyllous fungus – Miocene/Pliocene – Mycorrhizal fungus – North America – palaeoecology – taxonomy Introduction Fungal organic remains, usually fungal spores and dispersed sporocarps, are frequently found in a routine palynological investigation (Elsik 1996). -

Fungal Planet Description Sheets: 716–784 By: P.W
Fungal Planet description sheets: 716–784 By: P.W. Crous, M.J. Wingfield, T.I. Burgess, G.E.St.J. Hardy, J. Gené, J. Guarro, I.G. Baseia, D. García, L.F.P. Gusmão, C.M. Souza-Motta, R. Thangavel, S. Adamčík, A. Barili, C.W. Barnes, J.D.P. Bezerra, J.J. Bordallo, J.F. Cano-Lira, R.J.V. de Oliveira, E. Ercole, V. Hubka, I. Iturrieta-González, A. Kubátová, M.P. Martín, P.-A. Moreau, A. Morte, M.E. Ordoñez, A. Rodríguez, A.M. Stchigel, A. Vizzini, J. Abdollahzadeh, V.P. Abreu, K. Adamčíková, G.M.R. Albuquerque, A.V. Alexandrova, E. Álvarez Duarte, C. Armstrong-Cho, S. Banniza, R.N. Barbosa, J.-M. Bellanger, J.L. Bezerra, T.S. Cabral, M. Caboň, E. Caicedo, T. Cantillo, A.J. Carnegie, L.T. Carmo, R.F. Castañeda-Ruiz, C.R. Clement, A. Čmoková, L.B. Conceição, R.H.S.F. Cruz, U. Damm, B.D.B. da Silva, G.A. da Silva, R.M.F. da Silva, A.L.C.M. de A. Santiago, L.F. de Oliveira, C.A.F. de Souza, F. Déniel, B. Dima, G. Dong, J. Edwards, C.R. Félix, J. Fournier, T.B. Gibertoni, K. Hosaka, T. Iturriaga, M. Jadan, J.-L. Jany, Ž. Jurjević, M. Kolařík, I. Kušan, M.F. Landell, T.R. Leite Cordeiro, D.X. Lima, M. Loizides, S. Luo, A.R. Machado, H. Madrid, O.M.C. Magalhães, P. Marinho, N. Matočec, A. Mešić, A.N. Miller, O.V. Morozova, R.P. Neves, K. Nonaka, A. Nováková, N.H. -

Research Journal of Pharmaceutical, Biological and Chemical Sciences
ISSN: 0975-8585 Research Journal of Pharmaceutical, Biological and Chemical Sciences Phytochemical and Mineral Elements Composition of Bondazewia berkeleyi, Auricularia auricula and Ganoderma lucidum Fruiting Bodies. Emmanuel E Essien*, Victor N Mkpenie, and Stella M Akpan. Department of Chemistry, University of Uyo, Akwa Ibom State, Nigeria. ABSTRACT Fruiting bodies of wild edible medicinal mushrooms, Bondazewia berkeleyi, Auricularia auricula and Ganoderma lucidum, were analyzed for the presence of secondary metabolites and concentrations of toxic (Cd, Cr, Ni, Pb) and essential (Co, Cu, K, Li, Mn, Na, Zn) elements. The results revealed the presence of alkaloids, flavonoids, triterpenoids, saponins and carbohydrates in varied amounts. Tannins and phlobatannins were not detected. The levels (in ppm) of Na (156.80±310), K (246.20±6.62), Li (10.53±2.10), Zn (30.80±2.30), Cu (3.80±0.10), Mn (18.40±2.24), Co (2.98±0.17), Ni (0.024±0.080) and Cd (0.004±0.012) were highest in G. lucidum. Auricularia auricula showed the highest concentration (in ppm) of Pb (0.027±0.012) and Cr (0.005±0.100). However, the levels of the metals did not exceed the FAO/WHO stipulated dietary standards. This is the first chemical assessment of B. berkeleyi polypore. Keywords: Mushroom, Polypore, Secondary metabolites, Mineral nutrients, Dietary standards. *Corresponding author March – April 2015 RJPBCS 6(2) Page No. 200 ISSN: 0975-8585 INTRODUCTION Mushrooms are plant-like microorganisms, which grow like plant but are without chlorophyll. They depend on other organisms or plants for their nutrition. Information available in literature shows that mushrooms were first known to early Greeks and Romans who divided them into edible, poisonous, and medicinal mushrooms [1,2]. -

Mitochondrial Evolution in the Entomopathogenic Fungal Genus Beauveria
Received: 29 September 2020 | Revised: 12 October 2020 | Accepted: 13 October 2020 DOI: 10.1002/arch.21754 RESEARCH ARTICLE Mitochondrial evolution in the entomopathogenic fungal genus Beauveria Travis Glare1 | Matt Campbell2 | Patrick Biggs2 | David Winter2 | Abigail Durrant1 | Aimee McKinnon1 | Murray Cox2 1Bio‐Protection Research Centre, Lincoln University, Lincoln, New Zealand Abstract 2 School of Fundamental Sciences, Massey Species in the fungal genus Beauveria are pathogens of University, Palmerston North, New Zealand invertebrates and have been commonly used as the ac- Correspondence tive agent in biopesticides. After many decades with few Glare Travis, Bio‐Protection Research Centre, Lincoln University, PO Box 85084, Lincoln species described, recent molecular approaches to clas- 7647, New Zealand. sification have led to over 25 species now delimited. Email: [email protected] Little attention has been given to the mitochondrial genomes of Beauveria but better understanding may led to insights into the nature of species and evolution in this important genus. In this study, we sequenced the mi- tochondrial genomes of four new strains belonging to Beauveria bassiana, Beauveria caledonica and Beauveria malawiensis, and compared them to existing mitochon- drial sequences of related fungi. The mitochondrial gen- omes of Beauveria ranged widely from 28,806 to 44,135 base pairs, with intron insertions accounting for most size variation and up to 39% (B. malawiensis) of the mi- tochondrial length due to introns in genes. Gene order of the common mitochondrial genes did not vary among the Beauveria sequences, but variation was observed in the number of transfer ribonucleic acid genes. Although phylogenetic analysis using whole mitochondrial gen- omes showed, unsurprisingly, that B. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l.