Rapid Identification of Legionella Spp. by MALDI-TOF MS Based Protein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microbial Study on Corrosion
AN INVESTIGATION OF MICROBIAL DIVERSITY AND MICROBIOLOGICALLY INFLUENCED CORROSION IN AUTOMOTIVE FUEL ENVIRONMENTS by Charles H.D. Williamson IV A thesis submitted to the Faculty and the Board of Trustees of the Colorado School of Mines in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Environmental Science and Engineering). Golden, Colorado Date ____________________________ Signed: ___________________________ _ Charles H.D. Williamson IV Signed: ____________________________ Dr. John R. Spear Thesis Advisor Golden, Colorado Date ____________________________ Signed: ____________________________ Dr. John McCray Professor and Director Department of Civil and Environmental Engineering ii ABSTRACT Microbial contamination of fuels can cause issues such as biofouling, fuel degradation and microbiologically influenced corrosion (MIC). The focus of the research presented in this thesis was characterizing the microbial diversity of automotive fuels and automotive fuel environments in the United States via both molecular-based techniques as well as cultivation- based methods in order to gain insight into how this diversity is impacting fuels and fuel system infrastructure. A field survey of fuels including biodiesel, diesel, E10, E85, fuel-grade ethanol and gasoline was conducted; and 454 pyrosequencing of both 16S/18S rRNA genes as well as 16S/18S rRNA (transcribed into cDNA) was applied to identify both total and active microbial communities in these environments. Microbial communities in all fuel types were broadly similar, and prevalent phylotypes included Halomonas spp., Pseudomonas spp., Shewanella spp., Corynebacterium spp. and Acetobacter spp. Pyrosequencing libraries generated from cDNA and DNA indicated that the active and total communities of the sampled environments show significant overlap. The microbial communities of storage tanks containing fuel-grade ethanol and water were also characterized by molecular and cultivation-based techniques. -

NCTC) Bacterial Strain Equivalents to American Type Culture Collection (ATCC) Bacterial Strains
This list shows National Collection of Type Cultures (NCTC) bacterial strain equivalents to American Type Culture Collection (ATCC) bacterial strains. NCTC Number CurrentName ATCC Number NCTC 7212 Acetobacter pasteurianus ATCC 23761 NCTC 10138 Acholeplasma axanthum ATCC 25176 NCTC 10171 Acholeplasma equifetale ATCC 29724 NCTC 10128 Acholeplasma granularum ATCC 19168 NCTC 10172 Acholeplasma hippikon ATCC 29725 NCTC 10116 Acholeplasma laidlawii ATCC 23206 NCTC 10134 Acholeplasma modicum ATCC 29102 NCTC 10188 Acholeplasma morum ATCC 33211 NCTC 10150 Acholeplasma oculi ATCC 27350 NCTC 10198 Acholeplasma parvum ATCC 29892 NCTC 8582 Achromobacter denitrificans ATCC 15173 NCTC 10309 Achromobacter metalcaligenes ATCC 17910 NCTC 10807 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27061 NCTC 10808 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 17062 NCTC 10809 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27063 NCTC 12156 Acinetobacter baumannii ATCC 19606 NCTC 10303 Acinetobacter baumannii ATCC 17904 NCTC 7844 Acinetobacter calcoaceticus ATCC 15308 NCTC 12983 Acinetobacter calcoaceticus ATCC 23055 NCTC 8102 acinetobacter dna group 13 ATCC 17903 NCTC 10304 Acinetobacter genospecies 13 ATCC 17905 NCTC 10306 Acinetobacter haemolyticus ATCC 17907 NCTC 10305 Acinetobacter haemolyticus subsp haemolyticus ATCC 17906 NCTC 10308 Acinetobacter johnsonii ATCC 17909 NCTC 10307 Acinetobacter junii ATCC 17908 NCTC 5866 Acinetobacter lwoffii ATCC 15309 NCTC 12870 Actinobacillus delphinicola ATCC 700179 NCTC 8529 Actinobacillus equuli ATCC 19392 -

Legionella Gresilensis Sp. Nov. and Legionella Beliardensis Sp. Nov., Isolated from Water in France
International Journal of Systematic and Evolutionary Microbiology (2001), 51, 1949–1957 Printed in Great Britain Legionella gresilensis sp. nov. and Legionella beliardensis sp. nov., isolated from water in France 1 Centre National de Franc: ois Lo Presti,1‡ Serge Riffard,1 He! le' ne Meugnier,1 Re! fe! rence des Legionella 1 2 3 UPRES EA1655, Faculte! de Monique Reyrolle, Yves Lasne, † Patrick A. D. Grimont, Me! decine RTH Laennec, Francine Grimont,3 Robert F. Benson,4 Don J. Brenner,4 Rue Guillaume Paradin, 4 1 1 69372 Lyon cedex 08, Arnold G. Steigerwalt, Jerome Etienne and Jean Freney France 2 Laboratoire des Author for correspondence: Franc: ois Lo Presti. Tel: j33 0 169 79 79 60. Fax: j33 0 169 79 79 20. Radioisotopes et de e-mail: Francois.Lo-Presti!sanofi-synthelabo.com Biochimie mole! culaire, Ho# pital Edouard Herriot, ! ! 69437 Lyon cedex 03, Novel Legionella-like isolates, strains Montbeliard A1T and Greoux 11 D13T, France isolated from two different French water sources, were studied taxonomically 3 Unite! des Ente! robacte! ries, and phylogenetically. Morphological and biochemical characterization revealed Institut Pasteur, 75724 Paris cedex 15, France that they were Gram-negative, aerobic, non-spore-forming bacilli with a cut- glass appearance that grew only on L-cysteine-supplemented buffered charcoal 4 Respiratory Diseases Branch, Meningitis and yeast extract agar. Phenotypic characterization using fatty acid and ubiquinone Special Pathogens Branch, profiles and SDS-PAGE analysis confirmed that they were closely related, but National Center for distinct from, other species of the genus Legionella, since serotyping could not Infectious Diseases, Centers for Disease Control and relate them to any existing serogroup. -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Which Organisms Are Used for Anti-Biofouling Studies
Table S1. Semi-systematic review raw data answering: Which organisms are used for anti-biofouling studies? Antifoulant Method Organism(s) Model Bacteria Type of Biofilm Source (Y if mentioned) Detection Method composite membranes E. coli ATCC25922 Y LIVE/DEAD baclight [1] stain S. aureus ATCC255923 composite membranes E. coli ATCC25922 Y colony counting [2] S. aureus RSKK 1009 graphene oxide Saccharomycetes colony counting [3] methyl p-hydroxybenzoate L. monocytogenes [4] potassium sorbate P. putida Y. enterocolitica A. hydrophila composite membranes E. coli Y FESEM [5] (unspecified/unique sample type) S. aureus (unspecified/unique sample type) K. pneumonia ATCC13883 P. aeruginosa BAA-1744 composite membranes E. coli Y SEM [6] (unspecified/unique sample type) S. aureus (unspecified/unique sample type) graphene oxide E. coli ATCC25922 Y colony counting [7] S. aureus ATCC9144 P. aeruginosa ATCCPAO1 composite membranes E. coli Y measuring flux [8] (unspecified/unique sample type) graphene oxide E. coli Y colony counting [9] (unspecified/unique SEM sample type) LIVE/DEAD baclight S. aureus stain (unspecified/unique sample type) modified membrane P. aeruginosa P60 Y DAPI [10] Bacillus sp. G-84 LIVE/DEAD baclight stain bacteriophages E. coli (K12) Y measuring flux [11] ATCC11303-B4 quorum quenching P. aeruginosa KCTC LIVE/DEAD baclight [12] 2513 stain modified membrane E. coli colony counting [13] (unspecified/unique colony counting sample type) measuring flux S. aureus (unspecified/unique sample type) modified membrane E. coli BW26437 Y measuring flux [14] graphene oxide Klebsiella colony counting [15] (unspecified/unique sample type) P. aeruginosa (unspecified/unique sample type) graphene oxide P. aeruginosa measuring flux [16] (unspecified/unique sample type) composite membranes E. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

Subcellular Location of Piscirickettsia Salmonis Heat Shock Protein 60 (Hsp60) Chaperone by Using Immunogold Labeling and Proteomic Analysis
microorganisms Article Subcellular Location of Piscirickettsia salmonis Heat Shock Protein 60 (Hsp60) Chaperone by Using Immunogold Labeling and Proteomic Analysis 1, 2,3, 4 5 Cristian Oliver y, Patricio Sánchez y , Karla Valenzuela , Mauricio Hernández , Juan Pablo Pontigo 3, Maria C. Rauch 3, Rafael A. Garduño 4,6 , Ruben Avendaño-Herrera 2,7,* and Alejandro J. Yáñez 2,8,* 1 Laboratorio de Inmunología y Estrés de Organismos Acuáticos, Instituto de Patología Animal, Facultad de Ciencias Veterinarias, Universidad Austral de Chile, Valdivia 5090000, Chile; [email protected] 2 Interdisciplinary Center for Aquaculture Research, (INCAR), Concepción 4070386, Chile; [email protected] 3 Instituto de Bioquímica y Microbiología, Facultad de Ciencias, Universidad Austral de Chile, Valdivia 5090000, Chile; [email protected] (J.P.P.); [email protected] (M.C.R.) 4 Microbiology and Immunology Department, Dalhousie University, Halifax, NS B3H 4R2, Canada; [email protected] (K.V.); [email protected] (R.A.G.) 5 Austral-OMICS, Faculty of Sciences, Universidad Austral de Chile, Valdivia 5090000, Chile; [email protected] 6 Canadian Food Inspection Agency, Dartmouth Laboratory, Dartmouth, NS B3B 1Y9, Canada 7 Universidad Andrés Bello, Laboratorio de Patología de Organismos Acuáticos y Biotecnología Acuícola, Facultad Ciencias de la Vida, Viña del Mar 2531015, Chile 8 Facultad de Ciencias, Universidad Austral de Chile, Valdivia 5090000, Chile * Correspondence: [email protected] (R.A.-H.); [email protected] (A.J.Y.) These authors contributed equally to this work. y Received: 12 November 2019; Accepted: 31 December 2019; Published: 15 January 2020 Abstract: Piscirickettsia salmonis is the causative bacterial agent of piscirickettsiosis, a systemic fish disease that significantly impacts the Chilean salmon industry. -

Investigation of Bacterial Community Composition and Abundance in a Lowland Arable Catchment
Investigation of bacterial community composition and abundance in a lowland arable catchment A thesis submitted to the School of Environmental Sciences of the University of East Anglia in partial fulfilment of the degree of Doctor of Philosophy By Ali Khalaf A. Albaggar 2014 © This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that no quotation from the thesis, nor any information derived therefrom, may be published without the author’s prior consent. Abstract This study aimed to characterise the bacterial community composition and abundance in the River Wensum in Norfolk using epifluorescence microscopy (EFM), automated ribosomal intergenic analysis (ARISA) and 454 pyrosequencing. It also aimed to determine the effects of spatial and temporal variations and environmental factors on bacterial community composition and abundance in this intensively farmed lowland catchment. The three techniques provided the same trends in bacterial community composition and abundance across the Wensum catchment. Total bacterial numbers ranged from 0.21 × 10 6 cells/mL to 5.34 × 10 6 cells/mL (mean = 1.1 × 10 6 cells/mL). The bacterial community composition and abundance showed significant differences between sites and times and were related to environmental parameters, with temperature and flow rate explaining most of the variation in bacterial community composition and abundance. Bacterial abundance increases as water moves downstream, while bacterial diversity decreases as water moves downstream. Some operational taxonomic units (OTUs) become commoner as the water moves downstream (3 rd and 4 th order streams). This presumably reflects the fact that these bacteria are actively growing in the river, and reducing the abundance of other taxa. -

Application of Multilocus Sequence Analysis (MLSA) for Accurate Identification of Legionella Spp. Isolated from Municipal Founta
The Journal of Microbiology (2012) Vol. 50, No. 1, pp. 127–136 DOI 10.1007/s12275-012-1243-1 Copyright ⓒ 2012, The Microbiological Society of Korea Application of Multilocus Sequence Analysis (MLSA) for Accurate Identification of Legionella spp. Isolated from Municipal Fountains in Chengdu, China, Based on 16S rRNA, mip, and rpoB Genes Wang Guan1, Ying Xu1,2, Da-li Chen1, Legionella (http://www.ncbi.nlm.nih.gov/Taxonomy, accessed 1 1 1,3 July 25, 2011). Among them, approximately 20 are involved Jia-nan Xu , Yu Tian , and Jian-ping Chen * in human diseases. Legionella infection is mainly caused by inhalation of contaminated aerosol, leading to flu-like Pon- 1 Department of Parasitology, West China School of Preclinical and Forensic tiac fever or the more serious legionellosis, also known as Medicine, Sichuan University, Chengdu 610041, Sichuan, P. R. China 2Department of Clinical Laboratories, the First Affiliated Hospital of Legionnaires’ disease (LD). The latter is a form of severe Chengdu Medical College, Chengdu, 610500, Sichuan, P. R. China pneumonia with a fatality rate that may reach 50% in im- 3Animal Disease Prevention and Food Safety Key Laboratory of Sichuan munocompromised patients (Fields et al., 2002; Yu et al., Province, Sichuan University, Chengdu 610064, Sichuan, P. R. China 2002). One species, Legionella pneumophila, is responsible for about 90% of LD cases. The other species are rarely (Received May 16, 2011 / Accepted August 31, 2011) pathogenic, although Legionella longbeachae accounts for about 30% of cases of LD in Australia and New Zealand Legionellosis (Legionnaires’ disease; LD) is a form of severe (Helbig et al., 2002; Newton et al., 2010). -

Nosocomial Legionnaires' Disease
Frontiers in Science 2012, 2(4): 62-75 DOI: 10.5923/j.fs.20120204.03 Nosocomial Legionnaires’ Disease: Risque and Prevention Jalila Tai1,2, Mohamed Nabil Benchekroun2, Mly Mustapha Ennaji2, Mariam Mekkour1, Nozha Cohen1,* 1Division de Microbiologie et d’hygiène des Produits de l’Environnement, Institut Pasteur du Maroc, Casablanca, 20360, Maroc 2Laboratoire de Biotechnologie, de l’Environnement et de la Santé, Faculté des Sciences et Techniques, Université Hassan II-Mohammedia, 146, Maroc Abstract In 1977, Fraser et al. described an outbreak of pneumonia among legionnaires attending a convention at a hotel in Philadelphia in 1976. Legionnaires’ disease (LD) can be nosocomial, community acquired or travel related. The incidence of hospital-acquired legionellosis appears to be increasing. Colonization of water systems by Legionella spp. is ubiquitous in hospitals throughout the world. The outbreak, which later became known as legionnaires’ disease, was caused by a new pleomorphic, faintly staining gram-negative bacillus, L. pneumophila, which was isolated at the Center for Disease Control from lung tissues of legionnaires who died. Risk assessment for this disease forms the basis for the institution of control measures. Detection and quantification of Legionella spp. in the environment, in particular in the hospital water distribution system is one of the cornerstones of risk assessment. This review summarizes the current state-of-the-art regarding these aspects and points out important areas which require further study. The environmental surveillance revealed that the centralized hot water distribution system of the hospital was colonized with Legionella. Methods of prevention of the organisms for eradication involved in hospital water systems. -

EVALUATION of the P45 MOBILE INTEGRATIVE ELEMENT and ITS ROLE IN
EVALUATION OF THE p45 MOBILE INTEGRATIVE ELEMENT AND ITS ROLE IN Legionella pneumophila VIRULENCE A Dissertation by LANETTE M. CHRISTENSEN Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Jeffrey D. Cirillo Committee Members, James Samuel Jon Skare Farida Sohrabji Head of Program, Warren Zimmer May 2018 Major Subject: Medical Sciences Copyright 2018 Lanette Christensen ABSTRACT Legionella pneumophila are aqueous environmental bacilli that live within protozoal species and cause a potentially fatal form of pneumonia called Legionnaires’ disease. Not all L. pneumophila strains have the same capacity to cause disease in humans. The majority of strains that cause clinically relevant Legionnaires’ disease harbor the p45 mobile integrative genomic element. Contribution of the p45 element to L. pneumophila virulence and ability to withstand environmental stress were addressed in this study. The L. pneumophila Philadelphia-1 (Phil-1) mobile integrative element, p45, was transferred into the attenuated strain Lp01 via conjugation, designating p45 an integrative conjugative element (ICE). The resulting trans-conjugate, Lp01+p45, was compared with strains Phil-1 and Lp01 to assess p45 in virulence using a guinea pig model infected via aerosol. The p45 element partially recovered the loss of virulence in Lp01 compared to that of Phil-1 evident in morbidity, mortality, and bacterial burden in the lungs at the time of death. This phenotype was accompanied by enhanced expression of type II interferon in the lungs and spleens 48 hours after infection, independent of bacterial burden. -
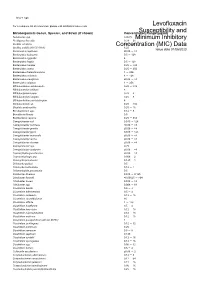
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 -