Proximate Composition and Fatty Acid Profile of South African Sardines (Sardinops Sagax Ocellatus) Using Conventional Techniques and Near Infrared (Nir) Spectroscopy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Sardinops Sagax Neopilchardus in Australia and New Zealand in 1995
DISEASES OF AQUATIC ORGANISMS Vol. 28: 1-16, 1997 Published January 16 Dis Aquat Org Epizootic mortality in the pilchard Sardinops sagax neopilchardus in Australia and New Zealand in 1995. I. Pathology and epizootiology 'New South Wales Agriculture, Elizabeth Macarthur Agricultural Institute, Private Bag 8, Camden, New South Wales 2570, Australia 'Fisheries Department of Western Australia, Animal Health Laboratory, 3 Baron-Hay Court, South Perth, Western Australia 6151. Australia 3National Institute of Water and Atmospheric Research, PO Box 14-901. Wellington, New Zealand "SIRO, Australian Animal Health Laboratory, PO Bag 24, Geelong, Victoria 3220, Australia ABSTRACT A large-scale eplzootic occurred In the Austialas~anpllchard Sard~nopssagax neo- p~lchardusbetween March and September 1995 ovei more than 5000 km of the Australian coastline and 500 km of the New Zealand coastline Affected fish died wlthln a tew mlnutes of cl~nicalslgns of respiratory distress and death was associated wlth hypoxaemla and hypercapnea Significant leslons were confined to the gllls and comprised acute to subacute inflammation followed by blzal re epithelia1 hypertrophy and hyperplasia The lesions were initially focal but progressed to become generalised over about 4 d Pathological changes in atfeded fish from xvestern Australia eastern Australia and New Zealand were simila~,suggesting a common aetiology The lesions were unllke those associated w~th ichthyotoxic algae, s~l~ceousalgae, phys~cochemicalfactors fungi, bacterla dinoflagellates, amoebde, other protozoa and inetazoa A herpesvlrus was consistently present In gills of affected flsh and absent from unaffected pilchards and IS proposed as the aetiological agent The late of spread of the mortal~ty front (approulmately 30 km d ') In relation to the migration late of pilchaids and plevaillng currents suggests that a vector was involved The dlsease may have been newly introduced lnto Australian wateis KEY WORDS Clupeoldel . -

Review of the Fishery Status for Whaler Sharks (Carcharhinus Spp.) in South Australian and Adjacent Waters
Review of the Fishery Status for Whaler Sharks (Carcharhinus spp.) in South Australian and adjacent waters Keith Jones FRDC Project Number 2004/067 January 2008 SARDI Aquatic Sciences Publication No. F2007/000721-1 SARDI Research Series No. 154 1 Review of the fishery status for whaler sharks in South Australian and adjacent waters. Final report to the Fisheries Research and Development Corporation. By: G.Keith Jones South Australian Research & Development Institute 2 Hamra Ave, West Beach SA 5022 (Current Address: PIRSA (Fisheries Policy) GPO Box 1625 Adelaide, SA 5001. Telephone: 08 82260439 Facsimile: 08 82262434 http://www.pirsa.saugov.sa.gov.au DISCLAIMER The author warrants that he has taken all reasonable care in producing this report. The report has been through the SARDI internal review process, and has been formally approved for release by the Chief Scientist. Although all reasonable efforts have been made to ensure quality, SARDI Aquatic Sciences does not warrant that the information in this report is free from errors or omissions. SARDI does not accept any liability for the contents of this report or for any consequences arising from its use or any other reliance placed upon it. © Copyright Fisheries Research and Development Corporation and South Australian Research & Development Institute, 2005.This work is copyright. Except as permitted under the Copyright Act 1968 (Commonwealth), no part of this publication may be reproduced by any process, electronic or otherwise, without the specific permission of the copyright owners. Neither may information be stored electronically in any form whatsoever without such permission. The Fisheries Research and Development Corporation plans, invests in and manages fisheries research and development throughout Australia. -

The White Shark (Carcharodon Carcharias) in the Ancient Peruvian Ceremonial Centre of Huaca Pucllana
International Journal of Osteoarchaeology Int. J. Osteoarchaeol. 26: 114–120 (2016) Published online 9 March 2014 in Wiley Online Library (wileyonlinelibrary.com) DOI: 10.1002/oa.2401 The White Shark (Carcharodon carcharias) in the Ancient Peruvian Ceremonial Centre of Huaca Pucllana A. ALTAMIRANO-SIERRAa* AND P. VARGAS-NALVARTEb a Áreas Costeras y Recursos Marinos (ACOREMA), Pisco, Peru b Museo de Sitio Huaca Pucllana, Calle General Borgoño cuadra 8 S/N, Lima 18, Peru ABSTRACT New data regarding the white shark (Carcharodon carcharias) at the archaeological complex Huaca Pucllana (200–700 AD) are presented on the basis of the recent discovery of teeth in ritual offering features. Previous information of this species from fossil, archaeological and modern records is reviewed. The use of the white sharks as an El Niño indicator is rejected. Past and present white shark distribution in the South East Pacificis reviewed, and the extermination of pinniped colonies as a factor in the poor modern record is discussed. Copyright © 2014 John Wiley & Sons, Ltd. Key words: Carcharodon carcharias; ENSO; Huaca Pucllana; Lima culture; palaeoecology; Peru Introduction investigated this culture include Julio C. Tello (1999), Pedro Villar Córdova (1935), Max Uhle The white shark (Carcharodon carcharias) is a large apex (1970), Thomas C. Patterson (1966) and Isabel Flores predator species distributed in most of the world’s (1981, 2005). Huaca Pucllana was mainly a village of oceans. Its habitat comprises coastal and offshore farmers and fishermen. Inhabitants of this site built waters of continental and insular shelves. This shark large adobe pyramids, where they worshiped deities species has a wide range of prey items, including birds, symbolized by figures associated with the sea and cetaceans, pinnipeds, osteichthyians, chondricthyians marine life (waves, sharks, sea lions, etc.). -

Shark Catch Trends and Effort Reduction in the Beach Protection Program, Kwazulu-Natal, South Africa (Elasmobranch Fisheries - Oral)
NOT TO BE CITED WITHOUT PRIOR REFERENCE TO THE AUTHOR(S) Northwest Atlantic Fisheries Organization Serial No. N4746 NAFO SCR Doc. 02/124 SCIENTIFIC COUNCIL MEETING – SEPTEMBER 2002 Shark Catch Trends and Effort Reduction in the Beach Protection Program, KwaZulu-Natal, South Africa (Elasmobranch Fisheries - Oral) S.F.J. Dudley Natal Sharks Board, P. Bag 2, Umhlanga Rocks, 4320, South Africa E-mail: [email protected] Abstract Shark nets have been set off the beaches of KwaZulu-Natal, South Africa, since 1952, to minimise risk of shark attack. Reliable catch data for each of the 14 shark species commonly caught are available from 1978 only. The nets fish in fixed localities very close to shore and there is an absence of fisheries independent data for most species. There is uncertainty about factors such as localised stock depletion and philopatry. Catch rates of seven species show a significant decline, but this figure drops to four with the exclusion of the confounding effects of the annual sardine run. Of the four, two are caught in very low numbers (Java Carcharhinus amboinensis and great hammerhead Sphyrna mokarran) and it is probable that any decline in population size reflects either local depletion or additional exploitation elsewhere. The other two species (blacktip C. limbatus and scalloped hammerhead S. lewini) are caught in greater numbers. C. limbatus appears to have been subject to local depletion. Newborn S. lewini are captured by prawn trawlers and discarded, mostly dead, adding to pressure on this species. As a precautionary measure, and in the absence of clarity on the question of stock depletion, in September 1999 a process of reducing the number of nets per installation was begun, with a view to reducing catches. -

Seafood Watch Report on the Pacific Sardine
Seafood Watch Seafood Report: Sardines Volume I image© Monterey Bay Aquarium Pacific sardine Sardinops sagax Alice Cascorbi Fisheries Research Analyst Monterey Bay Aquarium final 10 February 2004 Seafood Watch® Pacific Sardine Report February 10, 2004 About Seafood Watch® and the Seafood Reports Monterey Bay Aquarium’s Seafood Watch® program evaluates the ecological sustainability of wild-caught and farmed seafood commonly found in the United States marketplace. Seafood Watch® defines sustainable seafood as originating from sources, whether wild-caught or farmed, which can maintain or increase production in the long- term without jeopardizing the structure or function of affected ecosystems. Seafood Watch® makes its science-based recommendations available to the public in the form of regional pocket guides that can be downloaded from the Internet (seafoodwatch.org) or obtained from the Seafood Watch® program by emailing [email protected]. The program’s goals are to raise awareness of important ocean conservation issues and empower seafood consumers and businesses to make choices for healthy oceans. Each sustainability recommendation on the regional pocket guides is supported by a Seafood Report. Each report synthesizes and analyzes the most current ecological, fisheries and ecosystem science on a species, then evaluates this information against the program’s conservation ethic to arrive at a recommendation of “Best Choices”, “Good Alternatives” or “Avoid.” The detailed evaluation methodology is available upon request. In producing the Seafood Reports, Seafood Watch® seeks out research published in academic, peer-reviewed journals whenever possible. Other sources of information include government technical publications, fishery management plans and supporting documents, and other scientific reviews of ecological sustainability. -

Stable Isotope Analysis of Juvenile White Sharks Inside a Nursery Area Reveals Foraging in Demersal-Inshore Habitats and Trophic Overlap with Sympatric
fmars-08-687738 August 5, 2021 Time: 12:53 # 1 ORIGINAL RESEARCH published: 10 August 2021 doi: 10.3389/fmars.2021.687738 Stable Isotope Analysis of Juvenile White Sharks Inside a Nursery Area Reveals Foraging in Demersal-Inshore Habitats and Trophic Overlap With Sympatric Edited by: Sharks J. Marcus Drymon, 1† 1 † 1 † Mississippi State University, Emiliano García-Rodríguez , Sharon Z. Herzka * , Oscar Sosa-Nishizaki * , United States Christopher G. Lowe2 and John B. O’Sullivan3 Reviewed by: 1 Department of Biological Oceanography, Center for Scientific Research and Higher Education of Ensenada (CICESE), Simona Alessandra Ceriani, Ensenada, Mexico, 2 Department of Biological Sciences, California State University, Long Beach, CA, United States, Florida Fish and Wildlife Research 3 Monterey Bay Aquarium, Monterey, CA, United States Institute, United States Richard Reina, Monash University, Australia Knowledge about top predators’ trophic ecology is crucial for defining their role *Correspondence: in ecosystems, understanding habitat preferences, characterizing life stage-specific Sharon Z. Herzka feeding habits, and evaluating their interaction with fisheries. In the northeastern Pacific, [email protected] Oscar Sosa-Nishizaki white sharks (Carcharodon carcharias) occupy coastal habitats during the early life [email protected] stages, including Bahía Sebastián Vizcaíno (BSV) in Mexico, which is a known nursery †ORCID: area. Although BSV presumably provides high prey abundance, the trophic ecology of Emiliano García-Rodríguez orcid.org/0000-0002-9078-1300 immature white sharks is poorly understood. Carbon and nitrogen bulk stable isotope Sharon Z. Herzka analyses (SIA) were used to explore the trophic relationship of early life stages with orcid.org/0000-0001-7091-7656 their potential prey and to infer dietary overlap with sympatric sharks, while SIA of Oscar Sosa-Nishizaki orcid.org/0000-0002-3043-768X amino acids were used to estimate trophic position. -

Teleostei, Clupeiformes)
Old Dominion University ODU Digital Commons Biological Sciences Theses & Dissertations Biological Sciences Fall 2019 Global Conservation Status and Threat Patterns of the World’s Most Prominent Forage Fishes (Teleostei, Clupeiformes) Tiffany L. Birge Old Dominion University, [email protected] Follow this and additional works at: https://digitalcommons.odu.edu/biology_etds Part of the Biodiversity Commons, Biology Commons, Ecology and Evolutionary Biology Commons, and the Natural Resources and Conservation Commons Recommended Citation Birge, Tiffany L.. "Global Conservation Status and Threat Patterns of the World’s Most Prominent Forage Fishes (Teleostei, Clupeiformes)" (2019). Master of Science (MS), Thesis, Biological Sciences, Old Dominion University, DOI: 10.25777/8m64-bg07 https://digitalcommons.odu.edu/biology_etds/109 This Thesis is brought to you for free and open access by the Biological Sciences at ODU Digital Commons. It has been accepted for inclusion in Biological Sciences Theses & Dissertations by an authorized administrator of ODU Digital Commons. For more information, please contact [email protected]. GLOBAL CONSERVATION STATUS AND THREAT PATTERNS OF THE WORLD’S MOST PROMINENT FORAGE FISHES (TELEOSTEI, CLUPEIFORMES) by Tiffany L. Birge A.S. May 2014, Tidewater Community College B.S. May 2016, Old Dominion University A Thesis Submitted to the Faculty of Old Dominion University in Partial Fulfillment of the Requirements for the Degree of MASTER OF SCIENCE BIOLOGY OLD DOMINION UNIVERSITY December 2019 Approved by: Kent E. Carpenter (Advisor) Sara Maxwell (Member) Thomas Munroe (Member) ABSTRACT GLOBAL CONSERVATION STATUS AND THREAT PATTERNS OF THE WORLD’S MOST PROMINENT FORAGE FISHES (TELEOSTEI, CLUPEIFORMES) Tiffany L. Birge Old Dominion University, 2019 Advisor: Dr. Kent E. -

Diet Segregation Between Two Colonies of Little Penguins Eudyptula Minor in Southeast Australia
CORE Metadata, citation and similar papers at core.ac.uk Provided by Digital.CSIC Diet segregation between two colonies of little penguins Eudyptula minor in southeast Australia 1 2 3 ANDRÉ CHIARADIA, * MANUELA G. FORERO, KEITH A. HOBSON, 4 1,5 1 1 STEPHEN E. SWEARER, FIONA HUME, LEANNE RENWICK AND PETER DANN 1Research Department, Phillip Island Nature Parks, PO Box 97, Cowes,Vic. 3922, Australia (Email: [email protected]); 2Departamento de Biología de la Conser vación, Estación Biológica de Doñana, Avda Américo Vespucio, Sevilla, Spain; 3Environment Canada, Saskatoon, Saskatchewan, Canada; 4Department of Zoology, University of Melbourne, Melbourne,Victoria, and 5Private Bag 10, New Norfolk, Tasmania, Australia Abstract We studied foraging segregation between two different sized colonies of little penguins Eudyptula minor with overlapping foraging areas in pre-laying and incubation. We used stomach contents and stable isotope measurements of nitrogen (d15N) and carbon (d13C) in blood to examine differences in trophic position, prey-size and nutritional values between the two colonies. Diet of little penguins at St Kilda (small colony) relied heavily on anchovy while at Phillip Island (large colony), the diet was more diverse and anchovies were larger than those consumed by St Kilda penguins. Higher d15N values at St Kilda, differences in d13C values and the prey composition provided further evidence of diet segregation between colonies. Penguins from each colony took anchovies from different cohorts and probably different stocks, although these sites are only 70 km apart. Differences in diet were not reflected in protein levels in the blood of penguins, suggesting that variation in prey between colonies was not related to differences in nutritional value of the diet. -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -
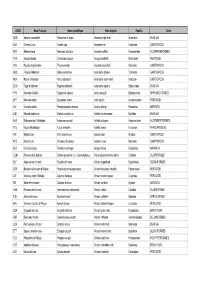
Liste Espèces
CODE Nom Français Nom scientifique Nom Anglais Famille Ordre ODQ Anomie cascabelle Pododesmus cepio Abalone jingle shell Anomiidae BIVALVIA ABX Ormeaux nca Haliotis spp Abalones nei Haliotidae GASTROPODA REN Sébaste rose Sebastes fasciatus Acadian redfish Scorpaenidae SCORPAENIFORMES YNA Acoupa toeroe Cynoscion acoupa Acoupa weakfish Sciaenidae PERCOIDEI HSV Pourpre aiguillonnee Thais aculeata Aculeate rock shell Muricidae GASTROPODA GBQ Troque d'Adanson Gibbula adansoni Adanson's gibbula Trochidae GASTROPODA NKA Natice d'Adanson Natica adansoni Adanson's moon snail Naticidae GASTROPODA GLW Tagal d'Adanson Tagelus adansonii Adanson's tagelus Solecurtidae BIVALVIA PYD Manchot d'Adélie Pygoscelis adeliae Adelie penguin Spheniscidae SPHENISCIFORMES QFT Maconde aden Synagrops adeni Aden splitfin Acropomatidae PERCOIDEI NIV Crevette adonis Parapenaeopsis venusta Adonis shrimp Penaeidae NATANTIA DJD Modiole adriatique Modiolus adriaticus Adriatic horse mussel Mytilidae BIVALVIA AAA Esturgeon de l'Adriatique Acipenser naccarii Adriatic sturgeon Acipenseridae ACIPENSERIFORMES FCV Fucus d'Adriatique Fucus virsoides Adriatic wrack Fucaceae PHAEOPHYCEAE IRR Mitre brûlée Mitra eremitarum Adusta miter Mitridae GASTROPODA KCE Murex bruni Chicoreus brunneus Adusta murex Muricidae GASTROPODA AES Crevette ésope Pandalus montagui Aesop shrimp Pandalidae NATANTIA CGM Poisson-chat, hybride Clarias gariepinus x C. macrocephalus Africa-bighead catfish, hybrid Clariidae SILURIFORMES SUF Ange de mer africain Squatina africana African angelshark Squatinidae SQUALIFORMES -

Bluefin Tuna
OCEAN SCIENCE SERIES Bluefin Tunas The State of the Science Executive Summary The three species of bluefin tunas can be found throughout the world’s oceans from the equator to sub-polar seas. A suite of physiological adaptations has allowed bluefin to range widely, exhibiting some of the greatest individual ranges of any fish. Some fisheries targeting bluefin tunas have been operating since ancient times, mainly supplying fish to small, local markets. Recent changes in the globalization of fish markets, coupled with industrial-scale fisheries and a high price in the global sushi market, have driven exploitation of bluefin tunas beyond sustainable levels. As a result, global populations have declined considerably, in some cases to as low as 3 percent of unfished population levels. The threat from overfishing is compounded by life history traits, such as slowness to reach maturity and a long life span, which means rebuilding depleted populations will be a lengthy process. Greater knowledge of the underlying biology of bluefin will allow scientists to understand how much fishing pressure is sustainable and how fast populations can recover. Ultimately, the recovery of bluefin tuna populations depends on the willingness of managers to enact scientifically sound management measures and on the ability of governments to enforce agreed upon rules. Bluefin Tunas: The State of the Science b Bluefin Tunas The State of the Science Andre Boustany, Ph.D.* Introduction the latest data on catch rates, as well as biologi- Among the world’s tuna species, none are as cal parameters of the species being assessed. large as or as individually valuable as the bluefin Species or populations that mature earlier gener- tunas. -

Teleostei, Clupeiformes) Early Larvae
Lat. Am. J. Aquat. Res., 42(1): 264-270, 2014 Neuromast development in larval clupeiforms 264 1 DOI: 103856/vol42-issue1-fulltext-22 Short Communication Development of free neuromasts in Engraulis ringens and Strangomera bentincki (Teleostei, Clupeiformes) early larvae Alejandra Llanos-Rivera1, Guillermo Herrera2, Eduardo Tarifeño3 & Leonardo R. Castro4 1Unidad de Biotecnología Marina, Facultad de Ciencias Naturales y Oceanográficas Universidad de Concepción, P.O. Box 160-C, Concepción, Chile 2Facultad de Ciencias, Universidad Católica de la Santísima Concepción Alonso de Rivera 2850, Concepción, Chile 3Laboratorio de Ecofisiología Marina, Departamento de Zoología, Facultad de Ciencias Naturales y Oceanográficas, Universidad de Concepción. P.O. Box 160-C, Concepción, Chile 4Laboratorio de Oceanografía Pesquera y Ecología Larval, Programa COPAS Sur Austral y Departamento de Oceanografía, Facultad de Ciencias Naturales y Oceanográficas Universidad de Concepción, P.O. Box 160-C, Concepción, Chile ABSTRACT. Neuromast morphology and distribution are characterized during early ontogeny of anchoveta (Engraulis ringens) and common sardine (Strangomera bentincki). Although both species share morphological features, they show several differences during their early ontogeny, such as size at hatching and yolk absorption. Larvae were obtained from incubation of planktonic eggs (at 12°C), collected during the spawning season 2001 (August-September) at Coliumo Bay. The neuromasts were observed from hatching to 25 days, and the pattern of neuromast appearance, in newly hatched larva, until yolk absorption, was determined using Janus Green staining and scanning electronic microscope. Results showed a similar pattern of neuromast development in both species. At hatching, two pairs of neuromasts were observed in the cephalic area and 8-9 in the rest of the body, which increased to 19 pairs and to 30-39 pairs at a larval size of 11 mm, respectively.