Rgraph2js: Usage from an R Session
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Windows 7 Operating Guide
Welcome to Windows 7 1 1 You told us what you wanted. We listened. This Windows® 7 Product Guide highlights the new and improved features that will help deliver the one thing you said you wanted the most: Your PC, simplified. 3 3 Contents INTRODUCTION TO WINDOWS 7 6 DESIGNING WINDOWS 7 8 Market Trends that Inspired Windows 7 9 WINDOWS 7 EDITIONS 10 Windows 7 Starter 11 Windows 7 Home Basic 11 Windows 7 Home Premium 12 Windows 7 Professional 12 Windows 7 Enterprise / Windows 7 Ultimate 13 Windows Anytime Upgrade 14 Microsoft Desktop Optimization Pack 14 Windows 7 Editions Comparison 15 GETTING STARTED WITH WINDOWS 7 16 Upgrading a PC to Windows 7 16 WHAT’S NEW IN WINDOWS 7 20 Top Features for You 20 Top Features for IT Professionals 22 Application and Device Compatibility 23 WINDOWS 7 FOR YOU 24 WINDOWS 7 FOR YOU: SIMPLIFIES EVERYDAY TASKS 28 Simple to Navigate 28 Easier to Find Things 35 Easy to Browse the Web 38 Easy to Connect PCs and Manage Devices 41 Easy to Communicate and Share 47 WINDOWS 7 FOR YOU: WORKS THE WAY YOU WANT 50 Speed, Reliability, and Responsiveness 50 More Secure 55 Compatible with You 62 Better Troubleshooting and Problem Solving 66 WINDOWS 7 FOR YOU: MAKES NEW THINGS POSSIBLE 70 Media the Way You Want It 70 Work Anywhere 81 New Ways to Engage 84 INTRODUCTION TO WINDOWS 7 6 WINDOWS 7 FOR IT PROFESSIONALS 88 DESIGNING WINDOWS 7 8 WINDOWS 7 FOR IT PROFESSIONALS: Market Trends that Inspired Windows 7 9 MAKE PEOPLE PRODUCTIVE ANYWHERE 92 WINDOWS 7 EDITIONS 10 Remove Barriers to Information 92 Windows 7 Starter 11 Access -

The Desktop (Overview)
The desktop (overview) The desktop is the main screen area that you see after you turn on your computer and log on to Windows. Like the top of an actual desk, it serves as a surface for your work. When you open programs or folders, they appear on the desktop. You can also put things on the desktop, such as files and folders, and arrange them however you want. The desktop is sometimes defined more broadly to include the taskbar and Windows Sidebar. The taskbar sits at the bottom of your screen. It shows you which programs are running and allows you to switch between them. It also contains the Start button , which you can use to access programs, folders, and computer settings. On the side of the screen, Sidebar contains small programs called gadgets. The desktop, taskbar, and Sidebar Where did my desktop go? Because programs run on top of the desktop, the desktop itself is often partially or completely hidden. But it's still there, underneath everything. To see the whole desktop without closing any of your open programs or windows, click the Show Desktop button on the taskbar. The desktop is revealed. Click the icon again to restore all of your windows to the way they were. Desktop Training Session Handout Page 1 http://ict.maxwell.syr.edu/vista/ Working with desktop icons Icons are small pictures that represent files, folders, programs, and other items. When you first start Windows, you'll see at least one icon on your desktop: the Recycle Bin (more on that later). -

Spot-Tracking Lens: a Zoomable User Interface for Animated Bubble Charts
Spot-Tracking Lens: A Zoomable User Interface for Animated Bubble Charts Yueqi Hu, Tom Polk, Jing Yang ∗ Ye Zhao y Shixia Liu z University of North Carolina at Charlotte Kent State University Tshinghua University Figure 1: A screenshot of the spot-tracking lens. The lens is following Belarus in the year 1995. Egypt, Syria, and Tunisia are automatically labeled since they move faster than Belarus. Ukraine and Russia are tracked. They are visible even when they go out of the spotlight. The color coding of countries is the same as in Gapminder[1], in which countries from the same geographic region share the same color. The world map on the top right corner provides a legend of the colors. ABSTRACT thus see more details. Zooming brings many benefits to visualiza- Zoomable user interfaces are widely used in static visualizations tion: it allows users to examine the context of an interesting object and have many benefits. However, they are not well supported in by zooming in the area where the object resides; labels overcrowded animated visualizations due to problems such as change blindness in the original view can be displayed without overlaps after zoom- and information overload. We propose the spot-tracking lens, a new ing in; it allows users to focus on a local area and thus reduce their zoomable user interface for animated bubble charts, to tackle these cognitive load. problems. It couples zooming with automatic panning and provides In spite of these benefits, zooming is not as well supported in an- a rich set of auxiliary techniques to enhance its effectiveness. -
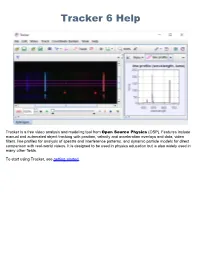
Tracker Helps You Mark the Steps but Does Not Limit Your Control Over Them
Tracker 6 Help Tracker is a free video analysis and modeling tool from Open Source Physics (OSP). Features include manual and automated object tracking with position, velocity and acceleration overlays and data, video filters, line profiles for analysis of spectra and interference patterns, and dynamic particle models for direct comparison with real-world videos. It is designed to be used in physics education but is also widely used in many other fields. To start using Tracker, see getting started. Getting Started When you first open Tracker it appears as shown below. Here's how to start analyzing a video: 1. Open a video or tracker file. 2. Identify the frames ("video clip") you wish to analyze. 3. Calibrate the video scale. 4. Set the reference frame origin and angle. 5. Track objects of interest with the mouse. 6. Plot and analyze the tracks. 7. Save your work in a tracker file. 8. Export track data to a spreadsheet. 9. Print, save or copy/paste images for reports. Note that the order of the buttons on the toolbar mirrors the steps used to analyze a video. For more information about Tracker's user interface, including user customization, see user interface. 1. Open a video or tracker file To open a local video, tracker tab (.trk), or tracker project (.trz) file, click the Open button or File|Open File menu item and select the file with the chooser, or drag and drop the file onto Tracker. You can also open still and animated image files (.jpg, .gif, .png), numbered sequences of image files, and images pasted from the clipboard. -

Digital Photo Frame User's Manual
PO_11960_XSU_00810B_Manual_en-gb Lang: English FI No.: 11960 Proof: 01 Date: 08/09/08 Page: 1 Digital Photo Frame User’s Manual Questions? Need some help? If you still have questions, call our Or visit help line found on the www.polaroid.com/support This manual should help you insert with this icon: understand your new product. PO_11960_XSU_00810B_Manual_en-gb Lang: English FI No.: 11960 Proof: 01 Date: 08/09/08 Page: 2 Controls and Basic Instructions CONGRATULATIONS on your purchase of a Polaroid Digital Photo Frame. Please read carefully and follow all instructions in the manual and those marked on the product before fi rst use. Failing to follow these warnings could result in personal injury or damage to the device. Also, remember to keep this User’s Manual in a convenient location for future reference. Important: Save the original box and all packing material for future shipping needs. Controls AUTO: Press it means exit.Hold for 2 seconds to start a Slide show. MENU: Press to enter or select. Hold for 2 seconds to enter the SETUP menu. UP ARROW Press to move up or select Previous. DOWN ARROW Press to move down or select Next. POWER: Turns power on or off. Installing a Flash Media Card USB Port 1. Find the slot that fi ts your fl ash media card. 2. Insert the card in the correct slot. 3. To remove the card, simply pull it out from the card slot. MS/SD/MMC Card Power Note: Do not remove any memory card from its slot while pictures are playing. -

1 Lecture 15: Animation
Lecture 15: Animation Fall 2005 6.831 UI Design and Implementation 1 1 UI Hall of Fame or Shame? Suggested by Ryan Damico Fall 2005 6.831 UI Design and Implementation 2 Today’s candidate for the Hall of Shame is this entry form from the 1800Flowers web site. The purpose of the form is to enter a message for a greeting card that will accompany a delivered flower arrangement. Let’s do a little heuristic evaluation of this form: Major: The 210 character limit is well justified, but hard for a user to check. Suggest a dynamic %-done bar showing how much of the quota you’ve used. (error prevention, flexibility & efficiency) Major: special symbols like & is vague. What about asterisk and hyphen – are those special too? What am I allowed to use, exactly? Suggest highlighting illegal characters, or beeping and not allowing them to be inserted. (error prevention) Cosmetic: the underscores in the Greeting Type drop-down menu look like technical identifiers, and some even look mispelled because they’ve omitted other punctuation. Bosss_Day? (Heuristic: match the real world) Major: how does Greeting Type affect card? (visibility, help & documentation) Cosmetic: the To:, Message,: and From: captions are not likely to align with what the user types (aesthetic & minimalist design) 2 Today’s Topics • Design principles • Frame animation • Palette animation • Property animation • Pacing & path Fall 2005 6.831 UI Design and Implementation 3 Today we’re going to talk about using animation in graphical user interfaces. Some might say, based on bad experiences with the Web, that animation has no place in a usable interface. -

Prof-UIS Frame Features White Paper
Prof-UIS Frame Features White Paper Published: January 2005 FOSS Software, Inc. 151 Main St., Suite 3 Salem, NH 03079 Phones: (603) 894 6425, (603) 894 6427 Fax: (603) 251 0077 E-mail: [email protected] Technical Support Forum: http://prof-uis.com/forum.aspx E-mail: [email protected] Contents Introduction.............................................................................................................................................3 Why choose us?.....................................................................................................................................4 Set of Samples.....................................................................................................................................4 CHM Help.............................................................................................................................................4 Technical Support ................................................................................................................................4 Royalty Free Licensing.........................................................................................................................4 Excellent Value ....................................................................................................................................4 Prof-UIS Feature List..............................................................................................................................5 GUI themes......................................................................................................................................5 -
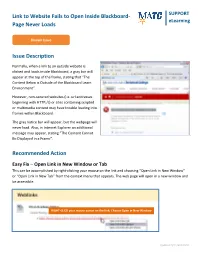
Link to Website Fails to Open Inside Blackboard- Page Never Loads Issue Description Recommended Action
Link to Website Fails to Open Inside Blackboard- SUPPORT eLearning Page Never Loads Known Issue Issue Description Normally, when a link to an outside website is clicked and loads inside Blackboard, a gray bar will appear at the top of the frame, stating that “The Content Below is Outside of the Blackboard Learn Environment”. However, non-secured websites (i.e. url addresses beginning with HTTP://) or sites containing scripted or multimedia content may have trouble loading into frames within Blackboard. The gray notice bar will appear, but the webpage will never load. Also, in Internet Explorer an additional message may appear, stating “The Content Cannot Be Displayed in a Frame”. Recommended Action Easy Fix – Open Link in New Window or Tab This can be accomplished by right-clicking your mouse on the link and choosing “Open Link in New Window” or “Open Link in New Tab” from the context menu that appears. The web page will open in a new window and be accessible. RIGHT-CLICK your mouse cursor on the link; Choose Open in New Window Updated 9/11/2013 MH Best Practice Fix – Create or Edit the Link to Open in New Window Automatically Links created through Blackboard’s Web link tool are easier to create and modify than links added to the text of a Blackboard Item. When creating or editing a link to your course, be sure to set the link to open in a new window. This will help ensure that the website is easily accessible in any browser going forwards. These instructions assume that you have EDIT MODE turned on in your course. -

01 Creation of HTML Pages with Frames, Links, Tables and Other Tags Date
www.vidyarthiplus.com Ex. No. : 01 Creation of HTML pages with frames, links, tables and other tags Date : AIM: To create a simple webpage using HTML that includes all tags. ALGORITHM: 1. Write a HTML program in the notepad with the tags such as A. FRAMES With frames, you can display more than one HTML document in the same browser window. Each HTML document is called a frame, and each frame is independent of the others. The Frameset Tag The <frameset> tag defines how to divide the window into frames. The Frame Tag The <frame> tag defines what HTML document to put into each frame. Example: <frameset cols="25%, 75 %"> <frame src="frame_a.htm"> <frame src="frame_b.htm"> </frameset> Tags and their Description: <frameset> Defines a set of frames <frame> Defines a sub window (a frame) www.vidyarthiplus.com www.vidyarthiplus.com B. LINKS A hyperlink is a reference (an address) to a resource on the web. Example: <a href="http://www.w3schools.com/">Visit W3Schools!</a> The href Attribute The href attribute defines the link "address". The target Attribute The target attribute defines where the linked document will be opened. Tag and its Description: <a> Defines an anchor C. TABLES Tables are defined with the <table> tag. A table is divided into rows (with the <tr> tag), and each row is divided into data cells (with the <td> tag). The letters td stands for "table data," which is the content of a data cell. Example: <table border="1"> <tr> <td>Row 1, cell 1</td> <td>Row 1, cell 2</td> </tr> </table> Tags and their Description: <Table> Defines a table <th> Defines a table header <tr> Defines a table row <td> Defines a table cell 2. -

PRO-TECH Drawing Quick Start
PRO-TECH Drawing Quick Start This guide will get you started with the basics of PRO-TECH’s drawing functionality. Topics discussed in this article include: creating simple elevations, drawing guidelines, editing object properties, explanation of defaults, and manipulating projects and elevations. This overview is only intended to get you started using the drawing program, for advanced use, please contact EDSS for further training options. Quick Start Conventions Whenever this document instructs you to “click” on an object, it means to use the left mouse button; otherwise, the document will specify the right mouse button. Also, “click” means to click the mouse button once; otherwise, the text will instruct you to “double-click”. Bolded text, in the instructions, either matches text on the application or represents a keyword to remember. Open the Drawing Module Open the drawing module from PRO-TECH. Figure 1 below depicts the main interface of the drawing module and contains the tools needed to create an elevation. Figure 1 1. Position your mouse over the Project Toolbar’s buttons and notice the pop-up hints that describe each image button. Take a brief moment to hover your mouse over the buttons to become familiar with their descriptions. 2. Notice the Caption Bar at the top of the screen. If your caption bar, like the one above, doesn’t contain a project name, a project is not open. If a project is not open, follow the instructions in the next section, Opening a Project. Otherwise, skip the next section and proceed to the section labeled: Creating a Blank Elevation. -

Desktop Studio: Hyperlinking Reports
Desktop Studio: Hyperlinking Reports Version: 18.1 Copyright © 2015 Intellicus Technologies This document and its content is copyrighted material of Intellicus Technologies. The content may not be copied or derived from, through any means, in parts or in whole, without a prior written permission from Intellicus Technologies. All other product names are believed to be registered trademarks of the respective companies. Dated: August 2015 Acknowledgements Intellicus acknowledges using of third-party libraries to extend support to the functionalities that they provide. For details, visit: http://www.intellicus.com/acknowledgements.htm 2 Contents 1 Making a field Hyperlink 4 Hyperlink the field to open a specific URL 4 3 1 Making a field Hyperlink When you make a component a clickable hyperlink, you can link a URL or a report with that report. Using this feature you can make "drill-down" reports or “linked reports”. Figure 1: Hyperlink options dialog box Hyperlink the field to open a specific URL 1. Click URL option button. 2. In the box provided below URL option button, specify the URL. 3. Click OK. 4 Hyperlink the field to open a specific report Figure 2: Report Parameters area of Hyperlink options dialog box 1. Click Drill down to another report option button. 2. In Select entry box, select the report that should be opened when the hyperlink is clicked. 3. Select the most appropriate option for Target. This is the way report in the hyperlink will open. 4. Specify Report Parameter and the value field if the report needs any report parameters to run. 5. Specify Save File Name to use when the report is implicitly published. -

Chapter 13 GUI Programming
Chapter 13 GUI Programming COSC 1436 Summer, 2018 July 19, 2018 Dr. Zhang Graphical User Interfaces • What is a GUI • –A graphical user interface allows the user to interact with the operating system and other programs using graphical elements such as icons, buttons, and dialog boxes. • Event-Driven • –GUI program must respond to the actions of the user. The user causes events to take place, such as the clicking of a button, and the program must respond to the events. • Using the tkinter Module The tkinter stands for “TK interface” Allows you to create simple GUI programs. Use import tkinter sentence Tkinter • The only GUI packaged with Python itself • Based on Tcl/Tk. Popular open-source scripting language/GUI widget set developed by John Ousterhout (90s) • Tk used in a wide variety of other languages (Perl, Ruby, PHP, etc.) • Cross-platform (Unix/Windows/MacOS) • It's small (~25 basic widgets) • http://effbot.org/tkinterbook/ Basic tkinter Widgets a component of an interface Typical steps in using Tkinter • You create and configure widgets (labels, buttons, sliders, etc.) • You pack them (geometry) • You implement functions that respond to various GUI events (event handling) • You run an event loop • Code example: SimpleGUI.py The Big Picture • A GUI lives in at least one graphical window • Here it is.... an empty window (no widgets) • This window is known as the "root" window • Usually only one root window per application • Code Example: empty_window.py Root(Main) Window • To create a new root window: • Import tkinter main_window=tkinter.Tk() • To start running the GUI, start its loop main_window.mainloop() • mainloop() function runs like an infinite loop until you close the main window.