We Hereby Approve the Dissertation Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Mapping Influenza-Induced Posttranslational Modifications On
viruses Article Mapping Influenza-Induced Posttranslational Modifications on Histones from CD8+ T Cells Svetlana Rezinciuc 1, Zhixin Tian 2, Si Wu 2, Shawna Hengel 2, Ljiljana Pasa-Tolic 2 and Heather S. Smallwood 1,3,* 1 Department of Pediatrics, University of Tennessee Health Science Center, Memphis, TN 38163, USA; [email protected] 2 Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA 99354, USA; [email protected] (Z.T.); [email protected] (S.W.); [email protected] (S.H.); [email protected] (L.P.-T.) 3 Children’s Foundation Research Institute, Memphis, TN 38105, USA * Correspondence: [email protected]; Tel.: +1-(901)-448–3068 Academic Editor: Italo Tempera Received: 10 October 2020; Accepted: 2 December 2020; Published: 8 December 2020 Abstract: T cell function is determined by transcriptional networks that are regulated by epigenetic programming via posttranslational modifications (PTMs) to histone proteins and DNA. Bottom-up mass spectrometry (MS) can identify histone PTMs, whereas intact protein analysis by MS can detect species missed by bottom-up approaches. We used a novel approach of online two-dimensional liquid chromatography-tandem MS with high-resolution reversed-phase liquid chromatography (RPLC), alternating electron transfer dissociation (ETD) and collision-induced dissociation (CID) on precursor ions to maximize fragmentation of uniquely modified species. The first online RPLC separation sorted histone families, then RPLC or weak cation exchange hydrophilic interaction liquid chromatography (WCX-HILIC) separated species heavily clad in PTMs. Tentative identifications were assigned by matching proteoform masses to predicted theoretical masses that were verified with tandem MS. We used this innovative approach for histone-intact protein PTM mapping (HiPTMap) to identify and quantify proteoforms purified from CD8 T cells after in vivo influenza infection. -

Child Neurology: Ethylmalonic Encephalopathy
Published Ahead of Print on February 28, 2020 as 10.1212/WNL.0000000000009144 RESIDENT & FELLOW SECTION Child Neurology: Ethylmalonic encephalopathy Periyasamy Govindaraj, PhD, Bindu Parayil Sankaran, FRACP, Madhu Nagappa, DM, Correspondence Hanumanthapura R. Arvinda, DM, Sekar Deepha, MSc, J.N. Jessiena Ponmalar, MSc, Sanjib Sinha, DM, Dr. Parayil Sankaran Narayanappa Gayathri, PhD, and Arun B. Taly, DM Bindu.parayilsankaran@ health.nsw.gov.au Neurology® 2020;94:e1-e4. doi:10.1212/WNL.0000000000009144 Ethylmalonic encephalopathy (EE; OMIM #602473) is an autosomal recessive disorder char- MORE ONLINE acterized by (1) progressive neurologic impairment, including global developmental delay with Video periods of regression during illness, progressive pyramidal and extrapyramidal signs, and seizures; and (2) generalized microvascular damage, including petechial purpura and chronic hemorrhagic diarrhea. It leads to premature death. EE is caused by mutations in ethylmalonic encephalopathy protein 1 (ETHE1), and more than 60 different mutations have been reported.1,2 ETHE1 encodes a mitochondrial sulfur dioxygenase involved in the catabolism of hydrogen sulfide 2 fi (H2S). Impairment of sulfur dioxygenase leads to the accumulation of hydrogen sul de and its fl derivatives (thiosulphate) in various body uids and tissues. Higher concentration of H2Sistoxic and induces direct damage to cell membranes. It inhibits cytochrome c oxidase (COX), increases lactic acid and short-chain acyl-COA dehydrogenase, and leads to elevation of ethyl malonate and C4/C5 acylcarnitine in muscle and brain.2 We describe the clinical phenotype and MRI, path- ologic, and biochemical findings of a patient with EE from India who had a novel homozygous c.493 G>C (p.D165H) variation in ETHE1. -

Exogenous Hydrogen Sulfide Plays an Important Role by Regulating
International Journal of Molecular Sciences Review Exogenous Hydrogen Sulfide Plays an Important Role by Regulating Autophagy in Diabetic-Related Diseases Shuangyu Lv , Huiyang Liu and Honggang Wang * Henan International Joint Laboratory of Nuclear Protein Regulation, School of Basic Medical Sciences, Henan University, Kaifeng 475000, China; [email protected] (S.L.); [email protected] (H.L.) * Correspondence: [email protected] Abstract: Autophagy is a vital cell mechanism which plays an important role in many physiological processes including clearing long-lived, accumulated and misfolded proteins, removing damaged organelles and regulating growth and aging. Autophagy also participates in a variety of biological functions, such as development, cell differentiation, resistance to pathogens and nutritional hunger. Recently, autophagy has been reported to be involved in diabetes, but the mechanism is not fully understood. Hydrogen sulfide (H2S) is a colorless, water-soluble, flammable gas with the typical odor of rotten eggs, which has been known as a highly toxic gas for many years. However, it has been reported recently that H2S, together with nitric oxide and carbon monoxide, is an important gas signal transduction molecule. H2S has been reported to play a protective role in many diabetes- related diseases, but the mechanism is not fully clear. Recent studies indicate that H2S plays an important role by regulating autophagy in many diseases including cancer, tissue fibrosis diseases and glycometabolic diseases; however, the related mechanism has not been fully studied. In this review, we summarize recent research on the role of H2S in regulating autophagy in diabetic-related diseases to provide references for future related research. -

Subcellular Distribution of HDAC1 in Neurotoxic Conditions Is Dependent on Serine Phosphorylation
The Journal of Neuroscience, August 2, 2017 • 37(31):7547–7559 • 7547 Neurobiology of Disease Subcellular Distribution of HDAC1 in Neurotoxic Conditions Is Dependent on Serine Phosphorylation Yunjiao Zhu,1* Oscar G. Vidaurre,1* XKadidia P. Adula,1 XNebojsa Kezunovic,1 XMaureen Wentling,1 X George W. Huntley,1 and XPatrizia Casaccia1,2 1Department of Neuroscience, Friedman Brain Institute, Graduate School of Biomedical Sciences, Icahn School of Medicine at Mount Sinai, New York, New York 10029, and 2Advanced Science Research Center at the Graduate Center of the City University of New York, New York, New York 10031 Calcium-dependent nuclear export of histone deacetylase 1 (HDAC1) was shown previously to precede axonal damage in culture, but the in vivo relevance of these findings and the potential posttranslational modifications of HDAC1 remained elusive. Using acute hippocam- pal slices from mice of either sex with genetic conditional ablation of Hdac1 in CA1 hippocampal neurons (i.e., Camk2a-cre;Hdac1 fl/fl), we show significantly diminished axonal damage in response to neurotoxic stimuli. The protective effect of Hdac1 ablation was detected also in CA3 neurons in Grik4-cre;Hdac1 fl/f mice, which were more resistant to the excitotoxic damage induced by intraventricular injection of kainic acid. The amino acid residues modulating HDAC1 subcellular localization were identified by site-directed mutagenesis, which identified serine residues 421 and 423 as critical for its nuclear localization. The physiological phosphorylation of HDAC1 was decreased by neurotoxic stimuli, which stimulated the phosphatase enzymatic activity of calcineurin. Treatment of neurons with the calcineurin inhibitors FK506 or cyclosporin A resulted in nuclear accumulation of phospho-HDAC1 and was neuroprotective. -

10.1136/Jmg.2005.036210 J Med Genet: First Published As 10.1136/Jmg.2005.036210 on 23 September 2005
JMG Online First, published on September 23, 2005 as 10.1136/jmg.2005.036210 J Med Genet: first published as 10.1136/jmg.2005.036210 on 23 September 2005. Downloaded from ETHE1 mutations are specific to Ethylmalonic Encephalopathy V. Tiranti1*, E. Briem 1*, E. Lamantea1, R. Mineri1, E. Papaleo2, L. De Gioia2, F. Forlani3, P. Rinaldo4, P. Dickson5, B. Abu-Libdeh6, L. Cindro-Heberle7, M. Owaidha8, R.M. Jack9, E. Christensen10, A. Burlina11, M. Zeviani1 1Unit of Molecular Neurogenetics – Pierfranco and Luisa Mariani Center for the Study of Children’s Mitochondrial Disorders, National Neurological Institute “C. Besta”, Milan, Italy; 2Department of Biotechnology and Biosciences. University of Milan-Bicocca, Italy; 3Dipartimento di Scienze Molecolari Agro-Alimentari, Facoltà di Agraria, State University of Milan, Italy; 4Laboratory Medicine and Pathology, Mayo Clinic and Foundation, Biochemical Genetics Laboratory, Rochester, MN, USA; 5Harbor-UCLA Medical Center, Division of Medical Genetics, Torrance, CA, USA; 6Makassed Hospital, Mount of Olives, Jerusalem, Israel; 7Paediatric Neurology Unit, Al Sabah Hospital, Kuwait; 8Al-Jahra Hospital, Kuwait; 9Biochemical Genetics lab, Children’s Hospital, Sand Point Way NE, Seattle, WA USA: 10Department of Clinical Genetics, Rigshospitalet, Copenhagen, Denmark; 11Division of Inborn Metabolic Diseases, Department of Pediatrics, University of Padua, Italy Correspondence to: Massimo Zeviani, MD, PhD Unit of Molecular Neurogenetics National Neurological Institute “Carlo Besta” Via Temolo 4, 20133 Milan, Italy FAX +39-02-23942619 Phone +39-02-23942630 E-mail: [email protected] [email protected] http://jmg.bmj.com/ URL: www.mitopedia.org *The first two authors should be regarded as joint First Authors. on September 24, 2021 by guest. -

Determining HDAC8 Substrate Specificity by Noah Ariel Wolfson A
Determining HDAC8 substrate specificity by Noah Ariel Wolfson A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biological Chemistry) in the University of Michigan 2014 Doctoral Committee: Professor Carol A. Fierke, Chair Professor Robert S. Fuller Professor Anna K. Mapp Associate Professor Patrick J. O’Brien Associate Professor Raymond C. Trievel Dedication My thesis is dedicated to all my family, mentors, and friends who made getting to this point possible. ii Table of Contents Dedication ....................................................................................................................................... ii List of Figures .............................................................................................................................. viii List of Tables .................................................................................................................................. x List of Appendices ......................................................................................................................... xi Abstract ......................................................................................................................................... xii Chapter 1 HDAC8 substrates: Histones and beyond ...................................................................... 1 Overview ..................................................................................................................................... 1 HDAC introduction -

Fruit Ripening and Storage
OPEN Citation: Horticulture Research (2014) 1, 6; doi:10.1038/hortres.2014.6 ß 2014 Nanjing Agricultural University All rights reserved 2052-7276/14 www.nature.com/hortres ARTICLE Dynamic changes in proteins during apple (Malus x domestica) fruit ripening and storage Yun Shi1, Li Jiang1, Li Zhang2, Ruoyi Kang1 and Zhifang Yu1 A proteomic study, using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight/time-of-flight, was conducted in apple fruit (cv. ‘Golden Delicious’) starting at 10 days prior to harvest through 50 days in storage. Total protein was extracted using a phenol/sodium dodecyl sulfate protocol. More than 400 protein spots were detected in each gel and 55 differentially expressed proteins (p,0.05) were subjected to matrix-assisted laser desorption/ionization time-of-flight/ time-of-flight analysis. Fifty-three of these proteins were finally identified using an apple expressed sequence tag database downloaded from Genome Database for Rosaceae and placed into six categories. The categories and the percentage of proteins placed in each category were stress response and defense (49.0%), energy and metabolism (34.0%), fruit ripening and senescence (5.6%), signal transduction (3.8%), cell structure (3.8%) and protein synthesis (3.8%). Proteins involved in several multiple metabolic pathways, including glycolysis, pentose–phosphate pathway, anti-oxidative systems, photosynthesis and cell wall synthesis, were downregulated, especially during the climacteric burst in respiration and during the senescent stages of fruit development. Proteins classified as allergens or involved in cell wall degradation were upregulated during the ripening process. Some protein spots exhibited a mixed pattern (increasing to maximal abundance followed by a decrease), such as 1-aminocyclopropane-1-carboxylate oxidase, L-ascorbate peroxidase and abscisic acid response proteins. -

Genome-Wide Analysis of Glyoxalase-Like Gene Families in Grape
Li et al. BMC Genomics (2019) 20:362 https://doi.org/10.1186/s12864-019-5733-y RESEARCHARTICLE Open Access Genome-wide analysis of glyoxalase-like gene families in grape (Vitis vinifera L.) and their expression profiling in response to downy mildew infection Tiemei Li1,2,3, Xin Cheng1,2,3, Yuting Wang1,2,3, Xiao Yin1,2,3, Zhiqian Li1,2,3, Ruiqi Liu1,2,3, Guotian Liu1,2,3, Yuejin Wang1,2,3 and Yan Xu1,2,3* Abstract Background: The glyoxalase system usually comprises two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII). This system converts cytotoxic methylglyoxal (MG) into non-toxic D-lactate in the presence of reduced glutathione (GSH) in two enzymatic steps. Recently, a novel type of glyoxalase III (GLYIII) activity has observed in Escherichia coli that can detoxify MG into D-lactate directly, in one step, without a cofactor. Investigation of the glyoxalase enzymes of a number of plant species shows the importance of their roles in response both to abiotic and to biotic stresses. Until now, glyoxalase gene families have been identified in the genomes of four plants, Arabidopsis, Oryza sativa, Glycine max and Medicago truncatula but no similar study has been done with the grapevine Vitis vinifera L. Results: In this study, four GLYI-like,twoGLYII-like and three GLYIII-like genesareidentifiedfromthegenomedatabaseof grape. All these genes were analysed in detail, including their chromosomal locations, phylogenetic relationships, exon-intron distributions, protein domain organisations and the presence of conserved binding sites. Using quantitative real-time PCR analysis (qRT-PCR), the expression profiles of these geneswereanalysedindifferent tissues of grape, and also when under infection stress from downy mildew (Plasmopara viticola). -

Up-Regulation of UVRAG by HDAC1 Inhibition Attenuates 5FU-Induced
ANTICANCER RESEARCH 38 : 271-277 (2018) doi:10.21873/anticanres.12218 Up-regulation of UVRAG by HDAC1 Inhibition Attenuates 5FU-induced Cell Death in HCT116 Colorectal Cancer Cells YOON KYUNG JO 1* , NA YEON PARK 1* , JI HYUN SHIN 1, DOO SIN JO 1, JI-EUN BAE 1, EUN SUN CHOI 1, SUNGHO MAENG 1, HONG BAE JEON 2, SEON AE ROH 3, JONG WOOK CHANG 4, JIN CHEON KIM 3,5 and DONG-HYUNG CHO 1 1Department of Gerontology, Graduate School of East-West Medical Science, Kyung Hee University, Seoul, Republic of Korea; 2Biomedical Research Institute, MEDIPOST Corporation, Seongnam, Republic of Korea; 3Asan Institute for Life Sciences, Asan Medical Center, Seoul, Republic of Korea; 4Stem Cell & Regenerative Medicine Institute, Samsung Medical Center, Seoul, Republic of Korea; 5Department of Surgery, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Republic of Korea Abstract. The ultraviolent irradiation resistance-associated Autophagy has important roles in cellular homeostasis gene (UVRAG), a component of the Beclin 1/autophagy- through the degradation of useless or damaged proteins and related 6 complex, regulates the autophagy initiation step organelles via lysosomes (1, 2). The primordial function of and functions in the DNA-damage response. UVRAG is autophagy is a response to stress, such as starvation, frequently mutated in various cancer types, and mutations of oxidative stress, and ion stress (1, 2). Given that multiple UVRAG increase sensitivity to chemotherapy by impairing signaling pathways are involved in the regulation of DNA-damage repair. In this study, we addressed the autophagy progress, various autophagy-related (ATG) epigenetic regulation of UVRAG in colorectal cancer cells. -
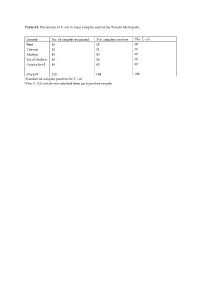
Supplementary File 1
Table S1. Prevalence of E. coli in meat samples sold at the Tamale Metropolis. Sample No. of samples examined aNo. samples positive bNo. E. coli Beef 45 39 39 Chevon 45 34 34 Mutton 45 40 40 Local chicken 45 36 36 Guinea fowl 45 40 40 Overall 225 189 189 aNumber of samples positive for E. coli. bOne E. Coli isolate was selected from each positive sample. Table S2. A table showing the eBURST (Based Upon Related Sequence Types) analyses of the study sequence types with global curated STs in Escherichia PubMLST database. MLST (Isolate) Type of clone Closet global ancestry Source sequence type (ST) ST69 (SG6) Similar a ST69 Animal (Food), Human ST155 (SLC2, Similar ST155 Animal (Food), Human, TLC13, CM4) Environment ST297 (TLC1) Similar ST297 Human ST1727 (NC3) Similar ST1727 Human ST44 (AC1) Single-Locus Variant ST10, ST752 Animal (Food), (SLV) b Human ST469 (CC6) Single-Locus Variant ST162 Food (SLV) ST540 (AB1, Single-Locus Variant ST4093 Human TG1) (SLV) ST1141 (NM11) Single-Locus Variant ST10, ST744 Animal (Food), (SLV) Human ST7473 (NB12) Single-Locus Variant ST10 Animal (Food), (SLV) Human ST6646 (CB1) Satellite c None - ST7483 (NB12) Satellite None - a Similar: study isolate was similar to a global curated known sequence type. b Single-Locus Variant (SLV): study isolate only shared similarity with global curated known sequence types that differed in one allelic gene. c Satellite: study isolate as a distantly related and did not shared any similarity with global curated known sequence types. Table S3. In silico identification and characterization of conserved stress response mechanisms in the E. -

Supplementary Information 2 to Accompany
1 Supplementary Information 2 to accompany 3 Sulfur-oxidizing symbionts without canonical genes for autotrophic CO2 fixation 4 Brandon K. B. Seah*1,7, Chakkiath Paul Antony1,8, Bruno Huettel2, Jan Zarzycki3, Lennart 5 Schada von Borzyskowski3, Tobias J. Erb3, Angela Kouris4, Manuel Kleiner5, Manuel 6 Liebeke1, Nicole Dubilier1,6, Harald R. Gruber-Vodicka1 7 1 Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany 8 2 Max Planck Genome Centre Cologne, Max Planck Institute for Plant Breeding Research, 9 Carl-von-Linné-Weg 10, 50829 Cologne, Germany 10 3 Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Str. 10, 35043 Marburg, 11 Germany 12 4 Energy Bioengineering and Geomicrobiology Group, University of Calgary, 2500 13 University Drive Northwest, Calgary, Alberta T2N 1N4, Canada 14 5 Department of Plant and Microbial Biology, North Carolina State University, Raleigh 15 27695, North Carolina, United States of America 16 6 MARUM, Center for Marine Environmental Sciences, University of Bremen, 28359 17 Bremen, Germany 18 7 Current address: Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 19 72076 Tübingen, Germany 20 8 Current address: Red Sea Research Center, Biological and Environmental Sciences and 21 Engineering (BESE) Division, King Abdullah University of Science and Technology 22 (KAUST), Thuwal 23955, Kingdom of Saudi Arabia 23 * Corresponding author 1 24 Supplementary Materials and Methods 25 Metabolite extraction and identification 26 Kentrophoros sp. H was collected on Elba in 2014 for metabolomics (Supplementary Table 27 7). Samples were fixed in 1 mL cold methanol (HPLC-grade, Sigma-Aldrich) and stored at - 28 20°C until use.