Viburnum Phylogeny Based on Combined Molecular Data: Implications for Taxonomy and Biogeography1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Toward a Resolution of Campanulid Phylogeny, with Special Reference to the Placement of Dipsacales
TAXON 57 (1) • February 2008: 53–65 Winkworth & al. • Campanulid phylogeny MOLECULAR PHYLOGENETICS Toward a resolution of Campanulid phylogeny, with special reference to the placement of Dipsacales Richard C. Winkworth1,2, Johannes Lundberg3 & Michael J. Donoghue4 1 Departamento de Botânica, Instituto de Biociências, Universidade de São Paulo, Caixa Postal 11461–CEP 05422-970, São Paulo, SP, Brazil. [email protected] (author for correspondence) 2 Current address: School of Biology, Chemistry, and Environmental Sciences, University of the South Pacific, Private Bag, Laucala Campus, Suva, Fiji 3 Department of Phanerogamic Botany, The Swedish Museum of Natural History, Box 50007, 104 05 Stockholm, Sweden 4 Department of Ecology & Evolutionary Biology and Peabody Museum of Natural History, Yale University, P.O. Box 208106, New Haven, Connecticut 06520-8106, U.S.A. Broad-scale phylogenetic analyses of the angiosperms and of the Asteridae have failed to confidently resolve relationships among the major lineages of the campanulid Asteridae (i.e., the euasterid II of APG II, 2003). To address this problem we assembled presently available sequences for a core set of 50 taxa, representing the diver- sity of the four largest lineages (Apiales, Aquifoliales, Asterales, Dipsacales) as well as the smaller “unplaced” groups (e.g., Bruniaceae, Paracryphiaceae, Columelliaceae). We constructed four data matrices for phylogenetic analysis: a chloroplast coding matrix (atpB, matK, ndhF, rbcL), a chloroplast non-coding matrix (rps16 intron, trnT-F region, trnV-atpE IGS), a combined chloroplast dataset (all seven chloroplast regions), and a combined genome matrix (seven chloroplast regions plus 18S and 26S rDNA). Bayesian analyses of these datasets using mixed substitution models produced often well-resolved and supported trees. -

Management of the Viburnum Leaf Beetle at the Morris Arboretum
University of Pennsylvania ScholarlyCommons Internship Program Reports Education and Visitor Experience 2017 Management of the Viburnum Leaf Beetle at the Morris Arboretum Anna Giesmann University of Pennsylvania Follow this and additional works at: https://repository.upenn.edu/morrisarboretum_internreports Part of the Botany Commons, and the Horticulture Commons Recommended Citation Giesmann, Anna, "Management of the Viburnum Leaf Beetle at the Morris Arboretum" (2017). Internship Program Reports. 10. https://repository.upenn.edu/morrisarboretum_internreports/10 An independent study project report by The John J. Willaman & Martha Haas Valentine Endowed Plant Protection Intern (2016-2017) This paper is posted at ScholarlyCommons. https://repository.upenn.edu/morrisarboretum_internreports/10 For more information, please contact [email protected]. Management of the Viburnum Leaf Beetle at the Morris Arboretum Abstract Pyrrhalta viburni (Coleoptera: Chrysomelidae), or the viburnum leaf beetle (VLB), is an invasive pest on viburnums in North America, where native species of the plant have little natural resistance. Resistance can be conferred by leaf texture, leaf chemistry, or a wound response that crushes VLB eggs. The beetle does not immediately kill host plants, but repeated defoliation is fatal after several years. Because viburnum is a common forest and landscape plant in the eastern United States, VLB is a serious concern. The Morris Arboretum has a large collection of viburnums, including many native and non-native species. While VLB had already been observed in passing, this project included a thorough baseline survey of VLB damage throughout the Arboretum. Data were collected for the number of twigs infested with VLB, the number of cavities on each twig, and whether a wound response had been produced. -

Nannyberry (Viburnum Lentago)
BWSR Featured Plant Name: Nannyberry (Viburnum lentago) Plant Family: Caprifoliaceae Statewide Wetland Indicator Status: Great Plains – FACU Nannyberry is a shrub that is both beautiful and Midwest – FAC versatile. This shade-tolerant, woody plant has flat N. Cent. N. East – FAC topped white flowers, fruit that is used by a wide variety of birds and wildlife, and vibrant fall color. Frequently used as a landscaping plant, it is successful as a tall barrier and wind break. Its ability to function as both a small tree and multi-stemmed shrub and ability to adapt to many environmental conditions makes it well suited to buffer planting and other soil stabilization projects. Flat-topped flower clusters and finely serrated leaves Identification This native shrub can be identified by its pointed buds, unique flowers, and fruit. Growing to around twelve feet tall in open habitats, the species commonly produces suckers and multiple stems. The newest branches are light green in color and glabrous and buds narrow to a point. With age, the branches become grey, scaly and rough. The egg-shaped leaves are simple and opposite with tips abruptly tapering to a sharp point. Leaf surfaces are shiny, dark green and hairless with reddish finely serrated margins. Fall color is a vibrant dark red. Numerous dense, flat-topped flower heads appear and bloom from May to June. Flowers are creamy white and bell to saucer-shaped. The flowers develop into elliptical berry-like drupes that turn a blue-black color from July to August. The Multi-stemmed growth form sweet and edible fruit is used by a variety of wildlife species but has a wet wool odor when decomposing, thus its alternate name – Sheepberry. -

A Guide to Selecting Landscape Plants for Wisconsin
A2865 A guide to selecting landscape plants for Wisconsin E.R. Hasselkus CONTENTS Deciduous trees tall, 2 medium, 4 low, 5 Evergreen trees, 7 Deciduous shrubs tall, 8 medium, 10 low, 11 Evergreen shrubs tall to medium, 13 low, 14 Vines, 14 Groundcovers, 15 Botanical names index, 17 Common names index, 19 A guide to selecting he following is a list of It is important to consider the site landscape plants plants recommended for requirements of each plant that you landscape use in select. Some plants are very exacting for Wisconsin Wisconsin. The list is not as to their preferences and will fail to exhaustive, but includes do well or may die in an unfavorable Tmost of the better ornamental plant location. Many plants are sensitive to species and cultivars (cultivated vari- poorly drained conditions. Use only eties) that are usually available for species tolerant of poor drainage in sale in the state. low, wet spots. Other species need a The plants listed vary widely as to well-drained, yet moist, soil. The “cool their height, growth habit or form, soil” requirement is met by soil that is color, texture, site and soil require- shaded or sloping toward the north. ments, and other characteristics. They Where shade is indicated in the adap- are grouped according to height cate- tation and remarks column, it refers to gories and a brief summary of each tolerance, not a requirement for shade. plant’s characteristics follows its Finally, be sure to choose plants that name. are hardy in your area. Wisconsin is When selecting plants from a list, one divided into six zones (see map) on often tends to consider the flower dis- the basis of minimum winter tempera- play first of all. -

Phylogeny and Phylogenetic Taxonomy of Dipsacales, with Special Reference to Sinadoxa and Tetradoxa (Adoxaceae)
PHYLOGENY AND PHYLOGENETIC TAXONOMY OF DIPSACALES, WITH SPECIAL REFERENCE TO SINADOXA AND TETRADOXA (ADOXACEAE) MICHAEL J. DONOGHUE,1 TORSTEN ERIKSSON,2 PATRICK A. REEVES,3 AND RICHARD G. OLMSTEAD 3 Abstract. To further clarify phylogenetic relationships within Dipsacales,we analyzed new and previously pub- lished rbcL sequences, alone and in combination with morphological data. We also examined relationships within Adoxaceae using rbcL and nuclear ribosomal internal transcribed spacer (ITS) sequences. We conclude from these analyses that Dipsacales comprise two major lineages:Adoxaceae and Caprifoliaceae (sensu Judd et al.,1994), which both contain elements of traditional Caprifoliaceae.Within Adoxaceae, the following relation- ships are strongly supported: (Viburnum (Sambucus (Sinadoxa (Tetradoxa, Adoxa)))). Combined analyses of C ap ri foliaceae yield the fo l l ow i n g : ( C ap ri folieae (Diervilleae (Linnaeeae (Morinaceae (Dipsacaceae (Triplostegia,Valerianaceae)))))). On the basis of these results we provide phylogenetic definitions for the names of several major clades. Within Adoxaceae, Adoxina refers to the clade including Sinadoxa, Tetradoxa, and Adoxa.This lineage is marked by herbaceous habit, reduction in the number of perianth parts,nectaries of mul- ticellular hairs on the perianth,and bifid stamens. The clade including Morinaceae,Valerianaceae, Triplostegia, and Dipsacaceae is here named Valerina. Probable synapomorphies include herbaceousness,presence of an epi- calyx (lost or modified in Valerianaceae), reduced endosperm,and distinctive chemistry, including production of monoterpenoids. The clade containing Valerina plus Linnaeeae we name Linnina. This lineage is distinguished by reduction to four (or fewer) stamens, by abortion of two of the three carpels,and possibly by supernumerary inflorescences bracts. Keywords: Adoxaceae, Caprifoliaceae, Dipsacales, ITS, morphological characters, phylogeny, phylogenetic taxonomy, phylogenetic nomenclature, rbcL, Sinadoxa, Tetradoxa. -

Viburnum L. Viburnum
VWYZ genera Layout 1/31/08 1:11 PM Page 1154 V Ericaceae—Heath family Vaccinium L. blueberry, cranberry Jason J. Griffin and Frank A. Blazich Dr. Griffin is assistant professor at Kansas State University’s Department of Horticulture, Forestry, and Recreation Resources, Manhatten, Kansas; Dr. Blazich is alumni distinguished graduate professor of plant propagation and tissue culture at North Carolina State University’s Department of Horticultural Science, Raleigh, North Carolina Occurrence, growth habit, and uses. There are about Although cranberry has been introduced successfully 150 to 450 species (the number varies by authority) of into cultivation in British Columbia, Washington, and deciduous or evergreen shrubs (rarely trees or vines) in Oregon, Wisconsin and Massachusetts remain the largest Vaccinium (Huxley 1992b; LHBH 1976; Vander Kloet producers; the crops for 2000 were estimated at 2.95 and 1988). The majority of species are native to North and South 1.64 million barrels, respectively (NASS 2001). America and eastern Asia (LHBH 1976; Vander Kloet Evergreen huckleberry grows along the Pacific Coast 1988). Some of the more commonly cultivated North and is valued for its attractive foliage, which is often used in American species are listed in table 1. Like other members flower arrangements (Everett 1981). Species of Vaccinium of the Ericaceae, Vaccinium species require an acidic (pH also are prized as landscape plants. Lowbush forms are used 4.0 to 5.2) soil that is moist, well drained, and high in organ- to form attractive ground covers or shrubs. Two cultivars of ic matter (3 to 15%). Symptoms of mineral nutrient defi- creeping blueberry (V. -

Ciliate Biodiversity and Phylogenetic Reconstruction Assessed by Multiple Molecular Markers Micah Dunthorn University of Massachusetts Amherst, [email protected]
University of Massachusetts Amherst ScholarWorks@UMass Amherst Open Access Dissertations 9-2009 Ciliate Biodiversity and Phylogenetic Reconstruction Assessed by Multiple Molecular Markers Micah Dunthorn University of Massachusetts Amherst, [email protected] Follow this and additional works at: https://scholarworks.umass.edu/open_access_dissertations Part of the Life Sciences Commons Recommended Citation Dunthorn, Micah, "Ciliate Biodiversity and Phylogenetic Reconstruction Assessed by Multiple Molecular Markers" (2009). Open Access Dissertations. 95. https://doi.org/10.7275/fyvd-rr19 https://scholarworks.umass.edu/open_access_dissertations/95 This Open Access Dissertation is brought to you for free and open access by ScholarWorks@UMass Amherst. It has been accepted for inclusion in Open Access Dissertations by an authorized administrator of ScholarWorks@UMass Amherst. For more information, please contact [email protected]. CILIATE BIODIVERSITY AND PHYLOGENETIC RECONSTRUCTION ASSESSED BY MULTIPLE MOLECULAR MARKERS A Dissertation Presented by MICAH DUNTHORN Submitted to the Graduate School of the University of Massachusetts Amherst in partial fulfillment of the requirements for the degree of Doctor of Philosophy September 2009 Organismic and Evolutionary Biology © Copyright by Micah Dunthorn 2009 All Rights Reserved CILIATE BIODIVERSITY AND PHYLOGENETIC RECONSTRUCTION ASSESSED BY MULTIPLE MOLECULAR MARKERS A Dissertation Presented By MICAH DUNTHORN Approved as to style and content by: _______________________________________ -

Nannyberry* Viburnum Lentago
Nannyberry* Viburnum lentago Height: 15 feet Spread: 10 feet Sunlight: Hardiness Zone: 2b Other Names: Sheepberry Nannyberry in bloom Description: Photo courtesy of NetPS Plant Finder A magnificent yet underutilized native shrub with many endearing qualities; large clusters of creamy white flowers in spring, showy blue fruit in fall and reliable fall color; an upright spreading plant, hardy and adaptable, can be grown as a small tree Ornamental Features Nannyberry features showy creamy white flat-top flowers at the ends of the branches in mid spring. The royal blue fruits are held in abundance in spectacular clusters from late summer to late winter. It has dark green foliage throughout the season. The glossy pointy leaves turn an outstanding brick red in the fall. Nannyberry flowers Landscape Attributes Photo courtesy of NetPS Plant Finder Nannyberry is a multi-stemmed deciduous shrub with an upright spreading habit of growth. Its average texture blends into the landscape, but can be balanced by one or two finer or coarser trees or shrubs for an effective composition. This shrub will require occasional maintenance and upkeep, and should only be pruned after flowering to avoid removing any of the current season's flowers. It is a good choice for attracting birds to your yard, but is not particularly attractive to deer who tend to leave it alone in favor of tastier treats. Gardeners should be aware of the following characteristic(s) that may warrant special consideration; - Suckering Nannyberry is recommended for the following landscape applications; Nannyberry fruit - Accent Photo courtesy of NetPS Plant Finder - Mass Planting - Hedges/Screening - General Garden Use - Naturalizing And Woodland Gardens Planting & Growing Nannyberry will grow to be about 15 feet tall at maturity, with a spread of 10 feet. -

Phylogeny and Phylogenetic Nomenclature of the Campanulidae Based on an Expanded Sample of Genes and Taxa
Systematic Botany (2010), 35(2): pp. 425–441 © Copyright 2010 by the American Society of Plant Taxonomists Phylogeny and Phylogenetic Nomenclature of the Campanulidae based on an Expanded Sample of Genes and Taxa David C. Tank 1,2,3 and Michael J. Donoghue 1 1 Peabody Museum of Natural History & Department of Ecology & Evolutionary Biology, Yale University, P. O. Box 208106, New Haven, Connecticut 06520 U. S. A. 2 Department of Forest Resources & Stillinger Herbarium, College of Natural Resources, University of Idaho, P. O. Box 441133, Moscow, Idaho 83844-1133 U. S. A. 3 Author for correspondence ( [email protected] ) Communicating Editor: Javier Francisco-Ortega Abstract— Previous attempts to resolve relationships among the primary lineages of Campanulidae (e.g. Apiales, Asterales, Dipsacales) have mostly been unconvincing, and the placement of a number of smaller groups (e.g. Bruniaceae, Columelliaceae, Escalloniaceae) remains uncertain. Here we build on a recent analysis of an incomplete data set that was assembled from the literature for a set of 50 campanulid taxa. To this data set we first added newly generated DNA sequence data for the same set of genes and taxa. Second, we sequenced three additional cpDNA coding regions (ca. 8,000 bp) for the same set of 50 campanulid taxa. Finally, we assembled the most comprehensive sample of cam- panulid diversity to date, including ca. 17,000 bp of cpDNA for 122 campanulid taxa and five outgroups. Simply filling in missing data in the 50-taxon data set (rendering it 94% complete) resulted in a topology that was similar to earlier studies, but with little additional resolution or confidence. -

Comparative Micromorphology and Anatomy of Flowers and Floral Secretory Structures in Two Viburnum Species
Protoplasma DOI 10.1007/s00709-016-0972-0 ORIGINAL ARTICLE Comparative micromorphology and anatomy of flowers and floral secretory structures in two Viburnum species Agata Konarska1 Received: 21 October 2015 /Accepted: 6 April 2016 # The Author(s) 2016. This article is published with open access at Springerlink.com Abstract In entomogamous plants, the presence and function Viburnum and the family Adoxaceae. Additionally, floral of floral secretory structures, whose main role is to attract and nectaries features are helpful in assessment of the related- pollinators, is strictly associated with the pollination ecology ness between taxa and provide better understanding of the and hence the reproductive success of the plant. The aims of floral biology and pollination ecology. the present paper were to analyse the micromorphology and anatomy of flower nectaries and stigmas in Viburnum opulus Keywords Histochemistry .Micromorphologyandanatomy . and V. lantana and to determine the function and microstruc- Nectaries and trichomes . Stigma microstructure . Viburnum ture of inflorescence trichomes in both taxa using light and flowering scanning electron microscopy as well as histochemical assays. It was found that stigmas were formed by papillae, which contained lipids, polysaccharides, tannins, and pigments. Introduction Stigmatic secretion proceeded via cuticular pores. Floral nec- taries formed a thick layer around the styles, and nectar was The genus Viburnum L. representing the family Adoxaceae secreted through numerous nectarostomata. There were no comprises approximately 160–200 species of evergreen, traces of vascular bundles penetrating the nectary tissue. In semi-evergreen, and deciduous shrubs or small trees growing turn, numerous tannin deposits were observed in the cells of primarily in the temperate climate zone of the northern hemi- the glandular parenchyma. -

Dipsacales Phylogeny Based on Chloroplast Dna Sequences
DIPSACALES PHYLOGENY BASED ON CHLOROPLAST DNA SEQUENCES CHARLES D. BELL,1, 2 ERIKA J. EDWARDS,1 SANG-TAE KIM,1 AND MICHAEL J. DONOGHUE 1 Abstract. Eight new rbcL DNA sequences and 15 new sequences from the 5' end of the chloroplast ndhF gene were obtained from representative Dipsacales and outgroup taxa. These were analyzed in combination with pre- viously published sequences for both regions. In addition, sequence data from the entire ndhF gene, the trnL-F intergenic spacer region,the trnL intron,the matK region, and the rbcL-atpB intergenic spacer region were col- lected for 30 taxa within Dipsacales. Phylogenetic relationships were inferred using maximum parsimony and maximum likelihood methods. Inferred tree topologies are in strong agreement with previous results from sep- arate and combined analyses of rbcL and morpholo gy, and confidence in most major clades is now very high. Concerning controversial issues, we conclude that Dipsacales in the traditional sense is a monophyletic group and that Triplostegia is more closely related to Dipsacaceae than it is to Valerianaceae. Heptacodium is only weakly supported as the sister group of the Caprifolieae (within which relationships remain largely unresolved), and the exact position of Diervilleae is uncertain. Within Morinaceae, Acanthocalyx is the sister group of Morina plus Cryptothladia. Dipsacales now provides excellent opportunities for comparative studies, but it will be important to check the congruence of chloroplast results with those based on data from other genomes. Keywords: Dipsacales, Adoxaceae, Caprifoliaceae, Morinaceae, Dipsacaceae, Valerianaceae, phylogeny, chloroplast DNA. The Dipsacales has traditionally included the Linnaea). It excludes Adoxa and its relatives, as C ap ri foliaceae (s e n s u l at o, i . -
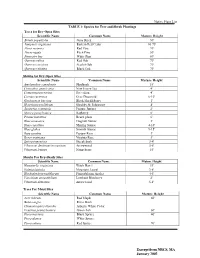
Native Plant List TABLE 1: Species for Tree and Shrub Plantings Trees For
Native Plant List TABLE 1: Species for Tree and Shrub Plantings Trees for Dry-Open Sites Scientific Name Common Name Mature Height Betula populifolia Gray Birch 30' Juniperis virginiana Eastern Red Cedar 10-75' Pinus resinosa Red Pine 70' Pin us rigida Pitch Pine 50' Pinus stro bus White Pine 80' Quercus rubra Red Oak 70' Quercus coccinea Scarlet Oak 70' Quercus velutina Black Oak 70' Shrubs for Dry-Open Sites Scientific Name 'Common Name Mature Height Amelanchier canadensis Shadbush 15' Ceanothus americanus New Jersey Tea 4' Comptonia peregrina Sweetfern 4' Cornus racemosa Gray Dogwood 6-10' Gaylussacia baccata Black Huckleberry l' Hypericum prolificum Shrubby St. Johnswort 4' Juniperus communis Pasture Juniper 2' Myrica pensylvanica Bayberry 6' Prunus maritima Beach plum 6' Rhus aromatica Fragrant Sumac 3' Rhus copallina Shining Sumac 4-10' Rhus glabra Smooth Sumac 9-15' Rosa carolina Pasture Rose 3' Rosa virginiana Virginia Rose 3' Spirea tomentosa Steeplebush 3-4' Viburnum dentatum/recognitum Arrowwood 5-8' Viburnum lentago Nannyberry 15' Shrubs For Dry-Shady Sites Scientific Name Common Name Mature Height Hamamelis wrginiana Witch Hazel 15' Kalmia latifolia Mountain Laurel 3-8' Rhododendron nudiflorum Pinxterbloom Azalea 4-6' Vaccinium angustifolium Lowbush Blueberry 2' Viburnum dentatum Arrowwood 5-8' Trees For Moist Sites Scientific Name Common Name Mature Height Acer rubrum Red Maple 60' Betula nigra River Birch Chamaecyparis thyoides Atlantic White Cedar Fraxinus pennsylvanica Green Ash 60' Picea mariana Black Spruce 40' Picea