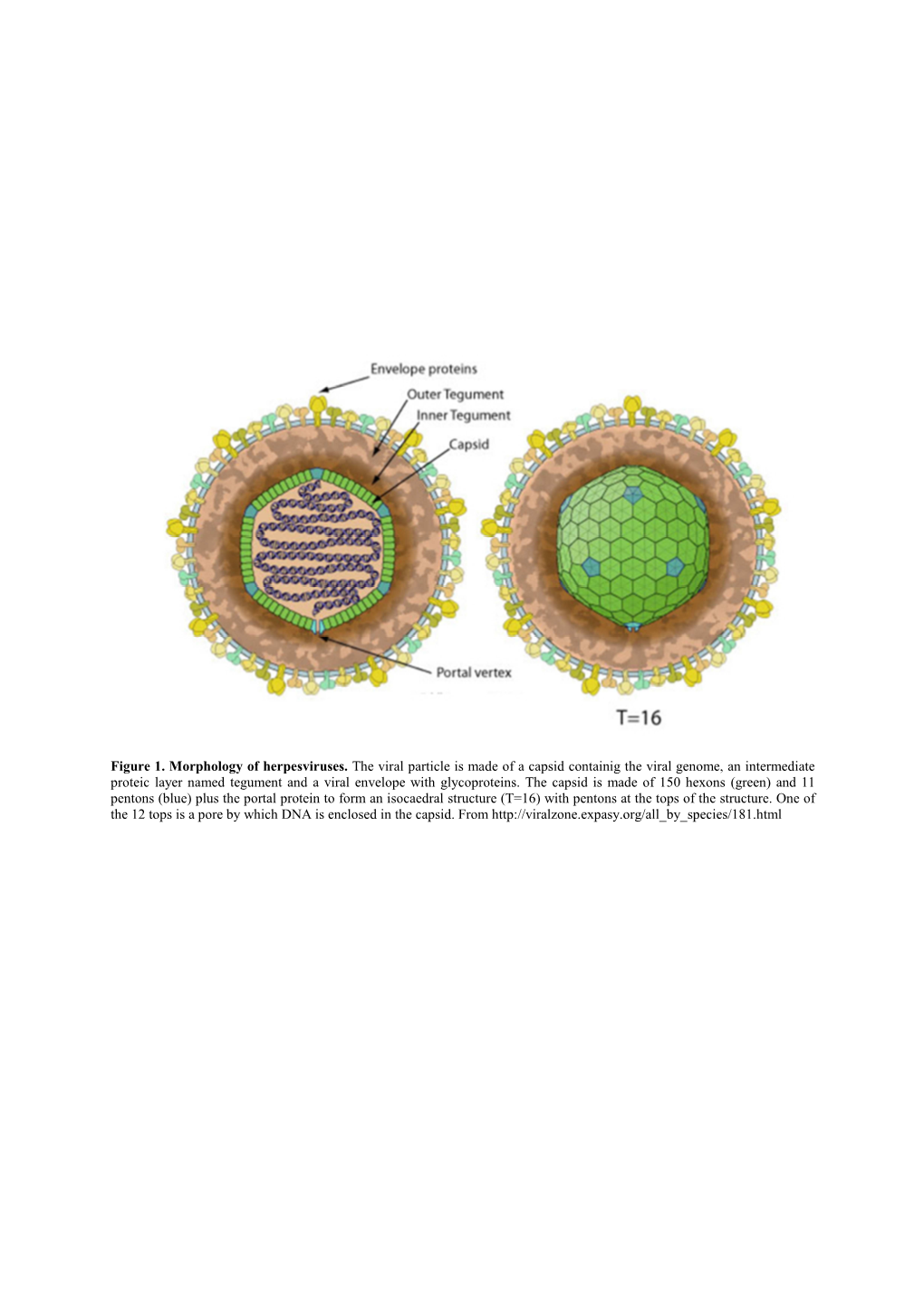
Figure 1. Morphology of Herpesviruses. the Viral Particle Is Made of A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Discovery of a Novel Bat Gammaherpesvirus
COMMENTARY Host-Microbe Biology crossmark Discovery of a Novel Bat Gammaherpesvirus Kurtis M. Host,a,b Blossom Damaniaa,b Lineberger Comprehensive Cancer Centera and Department of Microbiology and Immunology,b University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA ABSTRACT Zoonosis is the leading cause of emerging infectious diseases. In a re- cent article, R. S. Shabman et al. (mSphere 1[1]:e00070-15, 2016, 10.1128/ Published 17 February 2016 mSphere.00070-15) report the identification of a novel gammaherpesvirus in a cell Citation Host KM, Damania B. 2016. Discovery of a novel bat gammaherpesvirus. mSphere line derived from the microbat Myotis velifer incautus. This is the first report on a 1(1):e00016-16. doi:10.1128/mSphere.00016- replicating, infectious gammaherpesvirus from bats. The new virus is named bat 16. gammaherpesvirus 8 (BGHV8), also known as Myotis gammaherpesvirus 8, and is Copyright © 2016 Host and Damania. This is able to infect multiple cell lines, including those of human origin. Using next- an open-access article distributed under the terms of the Creative Commons Attribution 4.0 generation sequencing technology, the authors constructed a full-length annotated International license. genomic map of BGHV8. Phylogenetic analysis of several genes from BGHV8 re- Address correspondence to Blossom Damania, vealed similarity to several mammalian gammaherpesviruses, including Kaposi’s [email protected]. sarcoma-associated herpesvirus (KSHV). The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM. KEYWORDS: Myotis velifer incautus, bat, BGHV8, gammaherpesvirus, Myotis Discovery of a novel bat gammaherpesvirus 8 gammaherpesvirus merging infectious diseases (EID), a significant financial burden and public health Ethreat, are on the rise (1). -

Development of In-House Taqman Qpcr Assay to Detect Equine Herpesvirus-2 in Al-Qadisiyah City ﻟﺛﺎﻧﻲ ا ﻓﺎﯾرو
Iraqi Journal of Veterinary Sciences, Vol. 34, No. 2, 2020 (365-371) Development of in-house Taqman qPCR assay to detect equine herpesvirus-2 in Al-Qadisiyah city M.H. Al-Saadi Department of Internal and Preventive Medicine, College of Veterinary Medicine, University of Al-Qadisiyah, Al-Qadisiyah, Iraq, Email: [email protected] (Received September 6, 2019; Accepted October 1, 2019; Available online July 23, 2020) Abstract EHV-2 is distributed in horses globally. It is clustered within gamma-herpesvirus subfamily and percavirus genus. EHV-2 infection has two phases: latent and lytic. In the later, EHV-2 mainly associated with respiratory and genital symptoms. However, in the quiescent phase of infection, EHV-2 stay dormant in the host till viral reactivation. Our previous study has showed that EHV-2 can be harboured by equine tendons, suggesting that leukocytes possibly carrying EHV-2 for the systemic dissemination. So far, numerous PCR protocols have been performed targeting the gB gene. However, this gene is heterogenic. Therefore, there is a need to develop a quantitative diagnostic approach to detect the quiescent EHV-2 strains. To do this, Taqman qPCR assay was developed to quantify the virus. This was performed by targeting a highly conserved gene known as DNA polymerase (DPOL) gene using constructed plasmid as a standard curve calibrator. The obtained results showed an infection frequency of 33% in which the EHV-2 load reached 6647 copies/100 ng DNA whereas the minimum load revealed as 2 copies/100 ng DNA. The median quantification was found as 141 copies/ 100 ng DNA. -

Annual Conference 2016
Annual Conference 2016 POSTER ABSTRACT BOOK 21-24 MARCH 2016 ACC, LIVERPOOL, UK ANNUAL CONFERENCE 2016 SESSION 1 – MEMBRANE TRANSPORTERS S1/P1 the pump in this complex and it is conserved between bacterial species, with an average of 78.5% identity between the DNA Novel tripartite tricarboxylate transporters sequences and approximately 80% similarity between the amino acid sequences amongst Enterobacteriaceae. This pump acts as from Rhodopseudomonas palustris a drug-proton antiporter, four residues have been previously Leonardo Talachia Rosa, John Rafferty, reported as essential for proton translocation in Escherichia coli AcrB: D407, D408, K940 and T978. AcrB of E. coli has an identity David Kelly of 86% and a 94% similarity to that of S. Typhimurium. Based on The University of Sheffield, Sheffield, UK these data, we constructed an AcrB D408A chromosomal mutant in S. Typhimurium SL1344. Western blotting confirmed that the Rhodopseudomonas palustris is a soil non-sulfur purple mutant had the same level of expression of AcrB as the parental bacterium, with ability to degrade lignin-derived compounds and wild type strain. The mutant had no growth deficiencies either in also to generate high yields of hydrogen gas, what raises several LB or MOPS minimal media. However, compared with wild type biotechnological interests in this bacterium. Degradation SL1344, the mutant had decreased efflux activity and was pathways, though, must begin with substrate uptake. In this multi-drug hyper-susceptible. Interestingly, the phenotype of the context, Soluble Binding Proteins (SBP`s) dependant AcrB D408A mutant was almost identical to that of an ΔacrB transporters are responsible for high-affinity and specificity mutant. -

Prevalence and Risk Factors for Felis Catus Gammaherpesvirus 1
Veterinary Microbiology 238 (2019) 108426 Contents lists available at ScienceDirect Veterinary Microbiology journal homepage: www.elsevier.com/locate/vetmic Prevalence and risk factors for Felis catus gammaherpesvirus 1 detection in T domestic cats in Italy Francesca Caringellaa, Costantina Desarioa, Eleonora Lorussoa, Ivana Pallantea, Tommaso Furlanellob, Gianvito Lanavea, Gabriella Eliaa, Vito Martellaa, Roberta Iattaa, ⁎ Vanessa R. Barrsc, Julia Beattyc, Canio Buonavogliaa, Nicola Decaroa, a Department of Veterinary Medicine, University of Bari, Valenzano, Bari, Italy b San Marco Veterinary Clinic and Laboratory, Veggiano, Padova, Italy c Faculty of Science, Sydney School of Veterinary Science, The University of Sydney, Sydney, NSW, 2006, Australia ARTICLE INFO ABSTRACT Keywords: Felis catus gammaherpesvirus 1 (FcaGHV1), a novel gammaherpesvirus of domestic cats identified in 2014, has Cats been detected in different countries demonstrating a worldwide distribution. The aim of this study wastoes- Gammaherpesvirus tablish the prevalence of FcaGHV1 in Italy using a molecular epidemiological approach. FcaGHV1 DNA was Molecular survey detected with virus-specific real-time PCR in ≃1% of 2659 feline blood samples tested. Analysis of risk factors Risk factors showed that being male and coinfection with feline immunodeficiency virus (FIV) increase the likelihood of FcaGHV1 detection. One-third of FcaGHV1-positive cats also tested positive for FIV provirus, whereas coin- fections with feline panleukopenia virus were not demonstrated. Further studies are necessary to confirm the risk factors for FcaGHV1 detection and the pathobiology of the virus. 1. Introduction McLuckie et al., 2016a,b; Kurissio et al., 2018; Troyer et al., 2014), with detection rates between 9.6 and 23.6%. Herpesviridae are double-stranded DNA viruses (130-220 kbp) that Risk factors for FcaGHV1 infection are reported to be age, sex, comprise three subfamilies (Alpha-, Beta-, and Gammaherpesvirinae) on health status and coinfections with other microorganisms. -

Annual Conference 2018 Abstract Book
Annual Conference 2018 POSTER ABSTRACT BOOK 10–13 April, ICC Birmingham, UK @MicrobioSoc #Microbio18 Virology Workshop: Clinical Virology Zone A Presentations: Wednesday and Thursday evening P001 Rare and Imported Pathogens Lab (RIPL) turn around time (TAT) for the telephoned communication of positive Zika virus (ZIKV) PCR and serology results. Zaneeta Dhesi, Emma Aarons Rare and Imported Pathogens Lab, Public Health England, Salisbury, United Kingdom Abstract Background: RIPL introduced developmental assays for ZIKV PCR and serology on 18/01/16 and 10/03/16 respectively. The published ZIKV test TATs were 5 days for PCR and 7 days for serology. Methods: All ZIKV RNA positive, seroconversion and “probable” cases diagnosed at RIPL up until 31/05/17 were identified. For each case, the date on which the relevant positive sample was received, and the date on which it was telephoned out to the requestor was ascertained. The number of working days between these two dates was calculated. Results: ZIKV PCR - 151 ZIKV PCR positive results were identified, of which 4 samples were excluded because no TAT could be calculated. The mean TAT for the remaining 147 samples was 1.7 working days. Ninety percent of these results were telephoned within 3 or fewer days of the sample having been received. There was 1 sample where the TAT was above the 90th centile. ZIKV Serology - 147 seroconversion or “Probable” ZIKV cases diagnosed serologically were identified. The mean TAT for these samples was 2.5 working days. Ninety percent of these results were telephoned within 4 or fewer days of the sample having been received. -

The Critical Role of Genome Maintenance Proteins in Immune Evasion During Gammaherpesvirus Latency
fmicb-09-03315 January 4, 2019 Time: 17:18 # 1 REVIEW published: 09 January 2019 doi: 10.3389/fmicb.2018.03315 The Critical Role of Genome Maintenance Proteins in Immune Evasion During Gammaherpesvirus Latency Océane Sorel1,2 and Benjamin G. Dewals1* 1 Immunology-Vaccinology, Department of Infectious and Parasitic Diseases, Faculty of Veterinary Medicine-FARAH, University of Liège, Liège, Belgium, 2 Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, CA, United States Gammaherpesviruses are important pathogens that establish latent infection in their natural host for lifelong persistence. During latency, the viral genome persists in the nucleus of infected cells as a circular episomal element while the viral gene expression program is restricted to non-coding RNAs and a few latency proteins. Among these, the genome maintenance protein (GMP) is part of the small subset of genes expressed in latently infected cells. Despite sharing little peptidic sequence similarity, gammaherpesvirus GMPs have conserved functions playing essential roles in latent Edited by: Michael Nevels, infection. Among these functions, GMPs have acquired an intriguing capacity to evade University of St Andrews, the cytotoxic T cell response through self-limitation of MHC class I-restricted antigen United Kingdom presentation, further ensuring virus persistence in the infected host. In this review, we Reviewed by: Neil Blake, provide an updated overview of the main functions of gammaherpesvirus GMPs during University of Liverpool, latency with an emphasis on their immune evasion properties. United Kingdom James Craig Forrest, Keywords: herpesvirus, viral latency, genome maintenance protein, immune evasion, antigen presentation, viral University of Arkansas for Medical proteins Sciences, United States *Correspondence: Benjamin G. -

Lynx Canadensis)
bioRxiv preprint doi: https://doi.org/10.1101/579607; this version posted March 16, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Identification of a novel gammaherpesvirus in Canada lynx (Lynx canadensis) Liam D. Hendrikse1, Ankita Kambli1, Caroline Kayko2, Marta Canuti3, Bruce Rodrigues4, Brian Stevens5,6, Jennifer Vashon7, Andrew S. Lang3, David B. Needle5, Ryan M. Troyer1* 1Department of Microbiology and Immunology, University of Western Ontario, 1151 Richmond St., London, Ontario N6A 5C1, Canada 2Map and Data Centre, Western Libraries, University of Western Ontario, 1151 Richmond St., London, Ontario N6A 5C1, Canada 3Department of Biology, Memorial University of Newfoundland, 232 Elizabeth Ave., St. John's, Newfoundland and Labrador A1B 3X9, Canada 4Wildlife Division, Newfoundland and Labrador Department of Fisheries and Land Resources, P.O. Box 2007, Corner Brook, NL, A2H 7S1, Canada 5New Hampshire Veterinary Diagnostic Laboratory, College of Life Sciences and Agriculture, University of New Hampshire, Durham, New Hampshire, USA 6Canadian Wildlife Health Cooperative – Ontario/Nunavut, Guelph, Ontario, N1G 2W1, Canada 7Maine Department of Inland Fisheries and Wildlife, 650 State St., Bangor, Maine 04401, USA *author for correspondence: [email protected] Abstract Gammaherpesviruses (GHVs) infect many animal species and are associated with lymphoproliferative disorders in some. Previously, we identified several novel GHVs in North American felids, however a GHV had never been identified in Canada lynx (Lynx canadensis). We therefore hypothesized the existence of an unidentified GHV in lynx. -

Evidence to Support Safe Return to Clinical Practice by Oral Health Professionals in Canada During the COVID-19 Pandemic: a Repo
Evidence to support safe return to clinical practice by oral health professionals in Canada during the COVID-19 pandemic: A report prepared for the Office of the Chief Dental Officer of Canada. November 2020 update This evidence synthesis was prepared for the Office of the Chief Dental Officer, based on a comprehensive review under contract by the following: Paul Allison, Faculty of Dentistry, McGill University Raphael Freitas de Souza, Faculty of Dentistry, McGill University Lilian Aboud, Faculty of Dentistry, McGill University Martin Morris, Library, McGill University November 30th, 2020 1 Contents Page Introduction 3 Project goal and specific objectives 3 Methods used to identify and include relevant literature 4 Report structure 5 Summary of update report 5 Report results a) Which patients are at greater risk of the consequences of COVID-19 and so 7 consideration should be given to delaying elective in-person oral health care? b) What are the signs and symptoms of COVID-19 that oral health professionals 9 should screen for prior to providing in-person health care? c) What evidence exists to support patient scheduling, waiting and other non- treatment management measures for in-person oral health care? 10 d) What evidence exists to support the use of various forms of personal protective equipment (PPE) while providing in-person oral health care? 13 e) What evidence exists to support the decontamination and re-use of PPE? 15 f) What evidence exists concerning the provision of aerosol-generating 16 procedures (AGP) as part of in-person -

Diversification of Mammalian Deltaviruses by Host Shifting
Diversification of mammalian deltaviruses by host shifting Laura M. Bergnera,b,1, Richard J. Ortonb, Alice Broosb, Carlos Telloc,d, Daniel J. Beckere, Jorge E. Carreraf,g, Arvind H. Patelb, Roman Bieka, and Daniel G. Streickera,b,1 aInstitute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, Scotland; bMedical Research Center–University of Glasgow Centre for Virus Research, Glasgow G61 1QH, Scotland; cAssociation for the Conservation and Development of Natural Resources, 15037 Lima, Perú; dYunkawasi, 15049 Lima, Perú; eDepartment of Biology, University of Oklahoma, Norman, OK 73019; fDepartamento de Mastozoología, Museo de Historia Natural, Universidad Nacional Mayor de San Marcos, Lima 15081, Perú; and gPrograma de Conservación de Murciélagos de Perú, Piura 20001, Perú Edited by Paul E. Turner, Yale University, New Haven, CT, and approved November 25, 2020 (received for review September 22, 2020) Hepatitis delta virus (HDV) is an unusual RNA agent that replicates satellites either cospeciated with their hosts over ancient time- using host machinery but exploits hepatitis B virus (HBV) to scales or possess an unrecognized capacity for host shifting, mobilize its spread within and between hosts. In doing so, HDV which would imply their potential to emerge in novel species. enhances the virulence of HBV. How this seemingly improbable The latter scenario has been presumed unlikely since either both hyperparasitic lifestyle emerged is unknown, but it underpins the satellite and helper would need to be compatible with the novel likelihood that HDV and related deltaviruses may alter other host or deltaviruses would need to simultaneously switch host host–virus interactions. -

Investigation of Leporid Herpesvirus 4, an Emerging Pathogen of Rabbits: Infection and Prevalence Studies
Investigation of Leporid herpesvirus 4, an Emerging Pathogen of Rabbits: Infection and Prevalence Studies by Janet Ruth Sunohara-Neilson A Thesis presented to The University of Guelph In partial fulfilment of requirements for the degree of Doctor of Veterinary Science Guelph, Ontario, Canada © Janet R. Sunohara-Neilson, December, 2013 ABSTRACT INVESTIGATION OF LEPORID HERPESVIRUS 4, AN EMERGING PATHOGEN OF RABBITS: INFECTION AND PREVALENCE STUDIES Janet Sunohara-Neilson Advisor: University of Guelph, 2013 Dr. Patricia V. Turner Leporid herpesvirus 4 (LeHV-4) is a recently identified alphaherpesvirus that causes lethal respiratory disease in rabbits. Diagnosis has been dependent on the observation of distinctive intranuclear inclusion bodies in affected tissues. The objectives of this body of work were to describe the course of infection in laboratory rabbits, develop a serological test for the detection of antibodies to LeHV-4, and survey Ontario commercial meat rabbits and pet rabbits for LeHV- 4 antibody prevalence. Based on the results of an initial dose-range finding pilot study, 22 New Zealand white rabbits were inoculated intranasally with LeHV-4 and monitored for 22 days post- infection (dpi). Clinical signs of infection, including dyspnea, serous oculonasal discharge, pyrexia and weight loss, were evident from 2 to 7 dpi. LeHV-4 was isolated from nasal secretions between 2 and 10 dpi. Gross and microscopic pathology was evaluated and suppurative necrohemorrhagic pneumonia and splenic necrosis were the major findings at peak infection (5 to 7 dpi), at which time eosinophilic herpetic inclusions were present in nasal mucosa, skin, spleen, and lung. Virus neutralization (VN) assay demonstrated serum antibodies starting at 11 dpi and persisting until the study end (22 dpi). -

The Role of Viral Glycoproteins and Tegument Proteins in Herpes
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 2014 The Role of Viral Glycoproteins and Tegument Proteins in Herpes Simplex Virus Type 1 Cytoplasmic Virion Envelopment Dmitry Vladimirovich Chouljenko Louisiana State University and Agricultural and Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Veterinary Pathology and Pathobiology Commons Recommended Citation Chouljenko, Dmitry Vladimirovich, "The Role of Viral Glycoproteins and Tegument Proteins in Herpes Simplex Virus Type 1 Cytoplasmic Virion Envelopment" (2014). LSU Doctoral Dissertations. 4076. https://digitalcommons.lsu.edu/gradschool_dissertations/4076 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. THE ROLE OF VIRAL GLYCOPROTEINS AND TEGUMENT PROTEINS IN HERPES SIMPLEX VIRUS TYPE 1 CYTOPLASMIC VIRION ENVELOPMENT A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Interdepartmental Program in Veterinary Medical Sciences through the Department of Pathobiological Sciences by Dmitry V. Chouljenko B.Sc., Louisiana State University, 2006 August 2014 ACKNOWLEDGMENTS First and foremost, I would like to thank my parents for their unwavering support and for helping to cultivate in me from an early age a curiosity about the natural world that would directly lead to my interest in science. I would like to express my gratitude to all of the current and former members of the Kousoulas laboratory who provided valuable advice and insights during my tenure here, as well as the members of GeneLab for their assistance in DNA sequencing. -

Antibody Cross-Reactivity Between Porcine Cytomegalovirus (PCMV) and Human Herpesvirus-6 (HHV-6)
viruses Article Antibody Cross-Reactivity between Porcine Cytomegalovirus (PCMV) and Human Herpesvirus-6 (HHV-6) Uwe Fiebig 1, Angela Holzer 2, Daniel Ivanusic 1, Elena Plotzki 1, Hartmut Hengel 3 ID , Frank Neipel 2 and Joachim Denner 1,* ID 1 Robert Koch Institute, Nordufer 20, 13353 Berlin, Germany; [email protected] (U.F.); [email protected] (D.I.); [email protected] (E.P.) 2 Institute of Virology, University Erlangen, Schlossgarten 4, 91054 Erlangen, Germany; [email protected] (A.H.); [email protected] (F.N.) 3 Institute of Virology, Medical Center, Faculty of Medicine, University of Freiburg, Hermann-Herder-Strasse 11, 79104 Freiburg, Germany; [email protected] * Correspondence: [email protected]; Tel.: +49-30-18754-2800 Received: 25 July 2017; Accepted: 19 October 2017; Published: 28 October 2017 Abstract: Porcine cytomegalovirus (PCMV) infection is widely prevalent among pigs, and PCMV is one of the viruses which may be transmitted during xenotransplantation using pig cells, tissues, or organs. While human cytomegalovirus (HCMV) is a major risk factor for allotransplantation, it is still unclear whether PCMV is able to infect human cells or pose a risk for xenotransplantation. Previously, it was shown that transmission of PCMV after pig kidney to non-human primate transplantations resulted in a significantly reduced survival time of the transplanted organ. To detect PCMV, PCR-based and immunological methods were used. Screening of pigs by Western blot analyses using recombinant viral proteins revealed up to 100% of the tested animals to be infected. When the same method was applied to screen human sera for PCMV-reactive antibodies, positive Western blot results were obtained in butchers and workers in the meat industry as well as in normal blood donors.