Phylogenetic Analysis of Porcine Circovirus Type 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Henan IEE TA
Initial Environmental Examination August 2015 PRC: Henan Sustainable Livestock Farming and Product Safety Demonstration Project Prepared by the Henan Provincial Government for the Asian Development Bank CURRENCY EQUIVALENTS (as of 25 August 2015) Currency unit – yuan (CNY) CNY1.00 = $0.1562 $1.00 = CNY6.4040 ABBREVIATIONS ADB Asian Development Bank GHG greenhouse gas BOD5 5-day biochemical oxygen demand GRM grievance redress mechanism CNY Chinese yuan HPG Henan provincial government COD chemical oxygen demand IA implementing agency DO dissolved oxygen MOE Ministry of Environment EA executing agency PMO project management office EIA environmental impact assessment PPE project participating enterprise EIR environmental impact report RP resettlement plan EIT environmental impact table SOE state-owned enterprise EMP environmental management plan SPS Safeguard Policy Statement EPB environmental protection bureau WHO World Health Organization FSR feasibility study report WRB water resources bureau FYP five-year plan WTP water treatment plant GDP gross domestic product WWTP wastewater treatment plant WEIGHTS AND MEASURES oC degree centigrade m2 square meter dB decibel m3/a cubic meter per annum km kilometer m3/d cubic meter per day km2 square kilometer mg/kg milligram per kilogram kW kilowatt mg/l milligram per liter L liter mg/m3 milligram per cubic meter m meter t ton t/a ton per annum NOTE (i) In this report, "$" refers to US dollars. This initial environmental examination is a document of the borrower. The views expressed herein do not necessarily represent those of ADB's Board of Directors, Management, or staff, and may be preliminary in nature. Your attention is directed to the “terms of use” section of this website. -
Global Map of Irrigation Areas CHINA
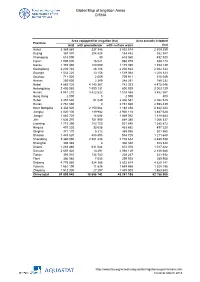
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52 -

Announcement of Interim Results for the Six Months Ended 30 June 2020
Hong Kong Exchanges and Clearing Limited and The Stock Exchange of Hong Kong Limited take no responsibility for the contents of this announcement, make no representation as to its accuracy or completeness and expressly disclaim any liability whatsoever for any loss howsoever arising from or in reliance upon the whole or any part of the contents of this announcement. (Stock Code: 0832) ANNOUNCEMENT OF INTERIM RESULTS FOR THE SIX MONTHS ENDED 30 JUNE 2020 FINANCIAL HIGHLIGHTS • Revenue for the six months ended 30 June 2020 amounted to RMB13,019 million, an increase of 43.6% compared with the corresponding period in 2019. • Gross profit margin for the period was 23.7%, a decrease of 3.6 percentage points compared with 27.3% for the corresponding period in 2019. • Profit attributable to equity shareholders of the Company for the period amounted to RMB727 million, an increase of 10.5% compared with the corresponding period in 2019. • Net profit margin for the period was 6.0%, a decrease of 2.5 percentage points compared with 8.5% for the corresponding period in 2019. • Basic earnings per share for the period was RMB26.43 cents, an increase of 9.8% compared with the corresponding period in 2019. • An interim dividend of HK11.0 cents per share for the six months ended 30 June 2020. 1 INTERIM RESULTS The board (the “Board”) of directors (the “Directors” and each a “Director”) of Central China Real Estate Limited (the “Company”) hereby announces the unaudited consolidated results of the Company and its subsidiaries (collectively, the “Group”) for -

China Report 2018 (FINAL – WEB)
CHINA’S CRACKDOWN ON CHRISTIANITY An analysis of the persecution of Christians in China in the first year of implementation of new Regulations for Religious Affairs (February 2018 – January 2019) A briefing document prepared for the Oireachtas Joint Committee on Foreign Affairs and Trade, and Defence (September 2019) TABLE OF CONTENTS Page 3 Introduction Pages 4 – 5 China Country Profile Page 6 Revised Regulations for Religious Affairs Page 7 Persecution of Other Religions Pages 8 – 10 Selected Case Histories Page 11 Map of China’s Provinces with Statistics Pages 12 – 33 List of Persecution Incidents (February 2018 – January 2019) Page 34 Conclusion Page 35 Recommendations Page 36 Sources COVER PHOTO Cross removed from church in Gongyi city in Henan Province in May 2018. (Photo Credit: Bitter Winter) Church in Chains is an independent Irish charity that encourages prayer and action in support of persecuted Christians worldwide. It is a member of the Department of Foreign Affairs and Trade Committee on Human Rights. This Briefing has been researched by Susanne Chipperfield (Operations Co-ordinator) and written by David Turner (Director). CHURCH IN CHAINS PO Box 10447, Glenageary, Co. Dublin, Ireland T 01-282 5393 E [email protected] W www.churchinchains.ie CHINA’s CRACKDOWN ON CHRISTIANITY Introduction and Background This briefing has been prepared by Church in Chains in response to the current crackdown on religion in China under the government’s “Sinicisation” policy – promoted by President Xi Jinping with the objective of creating a Chinese version of every religion and making every religion conform and be subservient to the Chinese Communist Party. -

Announcement of Interim Results for the Six Months Ended 30 June 2021
Hong Kong Exchanges and Clearing Limited and The Stock Exchange of Hong Kong Limited take no responsibility for the contents of this announcement, make no representation as to its accuracy or completeness and expressly disclaim any liability whatsoever for any loss howsoever arising from or in reliance upon the whole or any part of the contents of this announcement. (Stock Code: 0832) ANNOUNCEMENT OF INTERIM RESULTS FOR THE SIX MONTHS ENDED 30 JUNE 2021 FINANCIAL HIGHLIGHTS • Revenue for the six months ended 30 June 2021 amounted to RMB20,357 million, an increase of 56.4% compared with the corresponding period in 2020. • Gross profit margin for the period was 17.9%, a decrease of 5.8 percentage points compared with 23.7% for the corresponding period in 2020. • Profit attributable to equity shareholders of the Company for the period amounted to RMB729 million, an increase of 0.3% compared with the corresponding period in 2020. • Profit for the period was RMB1,025 million, representing an increase of 30.4% compared with the corresponding period in 2020. • Net profit margin for the period was 5.0%, a decrease of 1.0 percentage point compared with 6.0% for the corresponding period in 2020. • Basic earnings per share for the period was RMB25.63 cents, a decrease of 3.0% compared with the corresponding period in 2020. • Declaration of an interim dividend of HK14.75 cents per share for the six months ended 30 June 2021. 1 INTERIM RESULTS The board (the “Board”) of directors (the “Directors” and each a “Director”) of Central China Real Estate -

Distribution, Genetic Diversity and Population Structure of Aegilops Tauschii Coss. in Major Whea
Supplementary materials Title: Distribution, Genetic Diversity and Population Structure of Aegilops tauschii Coss. in Major Wheat Growing Regions in China Table S1. The geographic locations of 192 Aegilops tauschii Coss. populations used in the genetic diversity analysis. Population Location code Qianyuan Village Kongzhongguo Town Yancheng County Luohe City 1 Henan Privince Guandao Village Houzhen Town Liantian County Weinan City Shaanxi 2 Province Bawang Village Gushi Town Linwei County Weinan City Shaanxi Prov- 3 ince Su Village Jinchengban Town Hancheng County Weinan City Shaanxi 4 Province Dongwu Village Wenkou Town Daiyue County Taian City Shandong 5 Privince Shiwu Village Liuwang Town Ningyang County Taian City Shandong 6 Privince Hongmiao Village Chengguan Town Renping County Liaocheng City 7 Shandong Province Xiwang Village Liangjia Town Henjin County Yuncheng City Shanxi 8 Province Xiqu Village Gujiao Town Xinjiang County Yuncheng City Shanxi 9 Province Shishi Village Ganting Town Hongtong County Linfen City Shanxi 10 Province 11 Xin Village Sansi Town Nanhe County Xingtai City Hebei Province Beichangbao Village Caohe Town Xushui County Baoding City Hebei 12 Province Nanguan Village Longyao Town Longyap County Xingtai City Hebei 13 Province Didi Village Longyao Town Longyao County Xingtai City Hebei Prov- 14 ince 15 Beixingzhuang Town Xingtai County Xingtai City Hebei Province Donghan Village Heyang Town Nanhe County Xingtai City Hebei Prov- 16 ince 17 Yan Village Luyi Town Guantao County Handan City Hebei Province Shanqiao Village Liucun Town Yaodu District Linfen City Shanxi Prov- 18 ince Sabxiaoying Village Huqiao Town Hui County Xingxiang City Henan 19 Province 20 Fanzhong Village Gaosi Town Xiangcheng City Henan Province Agriculture 2021, 11, 311. -

Study on Hydrodynamic Water Quality Model and Dynamic Environmental Capacity of Qingyi River Basin
International Journal of Environmental Science and Development, Vol. 8, No. 6, June 2017 Study on Hydrodynamic Water Quality Model and Dynamic Environmental Capacity of Qingyi River Basin Yanpeng Wang, Li Wang, and Kuan Zhang other countries and regions because of its advanced and Abstract—Due to the change of natural conditions, the water reliability, the application research of EFDC model is being environmental capacity presenting time dynamic carried out in China in recently years. Yihui Chen Simulated characteristics. In this study, the Xuchang section of Qingyi the water dynamics and water quality in Dianchi Lake. The River Basin was taken as the research object, The EFDC model results of hydrodynamic simulation of the model are was adopted, COD and ammonia nitrogen as the main pollution factor, the dynamic water environmental capacity was studied. acceptable and close to the actual situation. Jianping Wang By constructing a one-dimensional hydrodynamic water quality combined with EFDC model, WASP model and GIS system model of Qinghe River Basin, Calculate of Water to study the Miyun Reservoir and its watershed nutrients, Environmental Capacity of Main Tributaries in Qingyi River which are beneficial for managing Miyun reservoir, Yixin Basin. The result shows that the environmental capacity of Yan studied the COD dynamics model of Yangtze River main stream is occupying the most proportion in the whole Estuary and got the distribution trend. river basin, Qingni River is much more than others in the main stream. In order to control the water quality of Qingyi River This study is based on the EFDC model, combined with Basin and control the total amount of pollutants in Qingyi River historical monitoring data and on-site mapping tools, analysis Basin, we established the water quality and water quality model hydrodynamic characteristics of river basins, using the in Qingyi River Basin. -

Minimum Wage Standards in China August 11, 2020
Minimum Wage Standards in China August 11, 2020 Contents Heilongjiang ................................................................................................................................................. 3 Jilin ............................................................................................................................................................... 3 Liaoning ........................................................................................................................................................ 4 Inner Mongolia Autonomous Region ........................................................................................................... 7 Beijing......................................................................................................................................................... 10 Hebei ........................................................................................................................................................... 11 Henan .......................................................................................................................................................... 13 Shandong .................................................................................................................................................... 14 Shanxi ......................................................................................................................................................... 16 Shaanxi ...................................................................................................................................................... -

Drilling Efficiency and Cost for Different Drill Technology in Loose Stratum
Drilling Efficiency and Cost for Different Drill Technology in Loose Stratum Ying Chen Doctoral student, Faculty of Engineering, China University of Geosciences, Wuhan, 430074, China e-mail: [email protected] Longchen Duan* Professor, Faculty of Engineering, China University of Geosciences, Wuhan, 430074, China *Corresponding Author, e-mail: [email protected] Yubei Lu Professor of engineering, Henan Engineering Research Center of Deep Exploration, Zhengzhou, 450053, China e-mail: [email protected] Ye Wu Professor, Henan Institute of Engineering, Zhengzhou, 451191, China e-mail: [email protected] ABSTRACT This study aims to analyze the drilling efficiency and cost in loosen stratum. Drilling technology includes mud positive circulation, pump-suction reverse circulation, and air downhole hammer. Drilling efficiency and cost data of 75 drilling projects in loosen stratum in Henan Province, China, were analyzed. Result shows the air downhole hammer has the highest drilling efficiency among drilling technologies in this study, and the average drilling efficiency is 3.76 and 2.17 times compared with mud positive and mud reverse circulation drilling technologies, respectively. The pump-suction reverse circulation drilling technology has the lowest cost in this study. The field study shows air downhole hammer drilling technology can be used in loosen stratum, and the cost still has reducing space. KEYWORDS: Drilling efficiency, drilling cost, mud positive circulation, pump- suction reverse circulation, air downhole hammer INTRODUCTION Drilling technology is widely used in geology exploration and engineering field, such as geotechnical engineering investigation, geological exploration, hydrological well, and water well drilling. Since drilling technology was invented, nowadays different types of drilling technology are being used in the drilling field. -
Huai River Basin Flood Management and Drainage Improvement Project
Document of The World Bank FOR OFFICIAL USE ONLY Public Disclosure Authorized Report No: 45437-CN PROJECT APPRAISAL DOCUMENT ON A PROPOSED LOAN Public Disclosure Authorized IN THE AMOUNT OF US$200 MILLION TO THE PEOPLE’S REPUBLIC OF CHINA FOR A HUAI RIVER BASIN FLOOD MANAGEMENT AND DRAINAGE IMPROVEMENT PROJECT Public Disclosure Authorized May 26, 2010 China and Mongolia Sustainable Development Unit Sustainable Development Department East Asia and Pacific Region Public Disclosure Authorized This document has a restricted distribution and may be used by recipients only in the performance of their official duties. Its contents may not otherwise be disclosed without World Bank authorization. CURRENCY EQUIVALENTS (Exchange Rate Effective February 2010) Currency Unit = Renminbi (RMB) Yuan (Y) Y1.0 = US$0.15 US$1.0 = Y6.827 FISCAL YEAR January 1 – December 31 ABBREVIATIONS AND ACRONYMS AEP Accelerated Emergency Program CNAO China National Audit Office CPCO Central Project Coordination Office CPLG Central Project Leading Group CPMO Central Project Management Office CPS Country Partnership Strategy DSR Dam Safety Report DUC Dam under Construction EG Expert Group EIA Environmental Impact Assessment EMP Environmental Management Plan FB Finance Bureau FDASS Flood Disaster Assessment and Support System FDIA Farmer Drainage and Irrigation Association HRBC Huai River Basin Commission MWR Ministry of Water Resources NDRC National Development and Reform Commission MIS Management Information System MOF Ministry of Finance O&M Operation and Maintenance PAP Project Affected People PAO Provincial Audit Office PDRC Provincial Development and Reform Commission PFB Provincial Finance Bureau PLG Project Leading Group PIU Project Implementation Unit POE Panel of Expert PPMO Provincial Project Management Office PWRB Provincial Water Resources Bureau RAP Resettlement Action Plan RO Resettlement Office SOCAD State Office for Comprehensive Agricultural Development Vice President: James W. -

2018 Interim Results Announcement
2018 Interim Results Announcement August 2018 Disclaimer DISCLAIMER This document has been prepared by Central China Real Estate Limited (the "Company") solely for use at this presentation held in connection with investor meetings and is subject to change without notice. The information contained in this presentation has not been independently verified. No representation or warranty, express or implied, is made and no reliance should be placed on the accuracy, fairness or completeness of the information presented. The Company, its affiliates, or any of their directors, officers, employees, advisers and representatives accept no liability whatsoever for any losses arising from any information contained in this presentation or otherwise arising in connection with this document. This presentation does not constitute or form part of, and should not be construed as, an offer to sell or a solicitation of an offer to buy any securities in the United States or any other jurisdictions in which such offer, solicitation or sale would be unlawful prior to registration or qualification under the securities laws of any such jurisdiction, and no part of this presentation shall form the basis of or be relied upon in connection with any contract or commitment. No securities may be offered or sold in the United States absent registration or an applicable exemption from registration requirements. Any public offering of securities to be made in the United States will be made by means of a prospectus. Such prospectus will contain detailed information about the Company making the offer and its management and financial statements. No public offering of any securities is to be made by the Company in the United States. -

A Spatial Econometric Perspective »
CORE Metadata, citation and similar papers at core.ac.uk Provided by Research Papers in Economics « A Fresh Scrutiny on Openness and Per Capita Income Spillovers in Chinese Cities: A Spatial Econometric Perspective » Selin OZYURT DR n°2008-17 A Fresh Scrutiny on Openness and Per Capita Income Spillovers in Chinese Cities: A Spatial Econometric Perspective Selin Ozyurt November 2008 Abstract This paper investigates openness and per capita income spillovers over 367 Chinese cities in the year 2004. Per capita income is modelled as dependent on investment, physical and social infrastructure, human capital, governmental policies and openness to the world. Our empirical analysis improves substantially the previous research in several respects: Firstly, by extending the data set to prefecture-level, it tackles the aggregation bias. Secondly, the introduction of recently developed explanatory spatial data analysis (ESDA) and spatial regression techniques allows to address misspecification issues due to spatial dependence. Thirdly, the endogeneity problem in the regression is taken into consideration through the use of generalised method of moments (GMM) estimator. Our major findings are in Chinese cities, physical and social infrastructure development, human capital and investment could be recognised as major driving sources of per capita income (i), whereas, the government expenditure ratio exerts a negative impact on per capita GDP level (ii). Our empirical findings also yield evidence on the existence of FDI and foreign trade spillovers in China (iii). These findings are robust to a number of alternative spatial weighting matrix specifications. Keywords: China, openness, spatial regression, spillovers, transition economies. JEL Classification: O11, O18, P24, R10. 1 1.