SUPPLEMENTAL DIGITAL CONTENT (SDC) 1 SDC-METHODS Microarray Processing: Total RNA Was Reverse-Transcribed Using the One-Cycle Ta
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nuclear and Mitochondrial Genome Defects in Autisms
UC Irvine UC Irvine Previously Published Works Title Nuclear and mitochondrial genome defects in autisms. Permalink https://escholarship.org/uc/item/8vq3278q Journal Annals of the New York Academy of Sciences, 1151(1) ISSN 0077-8923 Authors Smith, Moyra Spence, M Anne Flodman, Pamela Publication Date 2009 DOI 10.1111/j.1749-6632.2008.03571.x License https://creativecommons.org/licenses/by/4.0/ 4.0 Peer reviewed eScholarship.org Powered by the California Digital Library University of California THE YEAR IN HUMAN AND MEDICAL GENETICS 2009 Nuclear and Mitochondrial Genome Defects in Autisms Moyra Smith, M. Anne Spence, and Pamela Flodman Department of Pediatrics, University of California, Irvine, California In this review we will evaluate evidence that altered gene dosage and structure im- pacts neurodevelopment and neural connectivity through deleterious effects on synap- tic structure and function, and evidence that the latter are key contributors to the risk for autism. We will review information on alterations of structure of mitochondrial DNA and abnormal mitochondrial function in autism and indications that interactions of the nuclear and mitochondrial genomes may play a role in autism pathogenesis. In a final section we will present data derived using Affymetrixtm SNP 6.0 microar- ray analysis of DNA of a number of subjects and parents recruited to our autism spectrum disorders project. We include data on two sets of monozygotic twins. Col- lectively these data provide additional evidence of nuclear and mitochondrial genome imbalance in autism and evidence of specific candidate genes in autism. We present data on dosage changes in genes that map on the X chromosomes and the Y chro- mosome. -

Metallothionein Monoclonal Antibody, Clone N11-G
Metallothionein monoclonal antibody, clone N11-G Catalog # : MAB9787 規格 : [ 50 uL ] List All Specification Application Image Product Rabbit monoclonal antibody raised against synthetic peptide of MT1A, Western Blot (Recombinant protein) Description: MT1B, MT1E, MT1F, MT1G, MT1H, MT1IP, MT1L, MT1M, MT2A. Immunogen: A synthetic peptide corresponding to N-terminus of human MT1A, MT1B, MT1E, MT1F, MT1G, MT1H, MT1IP, MT1L, MT1M, MT2A. Host: Rabbit enlarge Reactivity: Human, Mouse Immunoprecipitation Form: Liquid Enzyme-linked Immunoabsorbent Assay Recommend Western Blot (1:1000) Usage: ELISA (1:5000-1:10000) The optimal working dilution should be determined by the end user. Storage Buffer: In 20 mM Tris-HCl, pH 8.0 (10 mg/mL BSA, 0.05% sodium azide) Storage Store at -20°C. Instruction: Note: This product contains sodium azide: a POISONOUS AND HAZARDOUS SUBSTANCE which should be handled by trained staff only. Datasheet: Download Applications Western Blot (Recombinant protein) Western blot analysis of recombinant Metallothionein protein with Metallothionein monoclonal antibody, clone N11-G (Cat # MAB9787). Lane 1: 1 ug. Lane 2: 3 ug. Lane 3: 5 ug. Immunoprecipitation Enzyme-linked Immunoabsorbent Assay ASSP5 MT1A MT1B MT1E MT1F MT1G MT1H MT1M MT1L MT1IP Page 1 of 5 2021/6/2 Gene Information Entrez GeneID: 4489 Protein P04731 (Gene ID : 4489);P07438 (Gene ID : 4490);P04732 (Gene ID : Accession#: 4493);P04733 (Gene ID : 4494);P13640 (Gene ID : 4495);P80294 (Gene ID : 4496);P80295 (Gene ID : 4496);Q8N339 (Gene ID : 4499);Q86YX0 (Gene ID : 4490);Q86YX5 -

Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation
From THE DEPARTMENT OF MICROBIOLOGY TUMOR AND CELL BIOLOGY (MTC) Karolinska Institutet, Stockholm, Sweden ROLE OF MITOCHONDRIAL RIBOSOMAL PROTEIN S18-2 IN CANCEROGENESIS AND IN REGULATION OF STEMNESS AND DIFFERENTIATION Muhammad Mushtaq Stockholm 2017 All previously published papers were reproduced with permission from the publisher. Published by Karolinska Institutet. Printed by E-Print AB 2017 © Muhammad Mushtaq, 2017 ISBN 978-91-7676-697-2 Role of Mitochondrial Ribosomal Protein S18-2 in Cancerogenesis and in Regulation of Stemness and Differentiation THESIS FOR DOCTORAL DEGREE (Ph.D.) By Muhammad Mushtaq Principal Supervisor: Faculty Opponent: Associate Professor Elena Kashuba Professor Pramod Kumar Srivastava Karolinska Institutet University of Connecticut Department of Microbiology Tumor and Cell Center for Immunotherapy of Cancer and Biology (MTC) Infectious Diseases Co-supervisor(s): Examination Board: Professor Sonia Lain Professor Ola Söderberg Karolinska Institutet Uppsala University Department of Microbiology Tumor and Cell Department of Immunology, Genetics and Biology (MTC) Pathology (IGP) Professor George Klein Professor Boris Zhivotovsky Karolinska Institutet Karolinska Institutet Department of Microbiology Tumor and Cell Institute of Environmental Medicine (IMM) Biology (MTC) Professor Lars-Gunnar Larsson Karolinska Institutet Department of Microbiology Tumor and Cell Biology (MTC) Dedicated to my parents ABSTRACT Mitochondria carry their own ribosomes (mitoribosomes) for the translation of mRNA encoded by mitochondrial DNA. The architecture of mitoribosomes is mainly composed of mitochondrial ribosomal proteins (MRPs), which are encoded by nuclear genomic DNA. Emerging experimental evidences reveal that several MRPs are multifunctional and they exhibit important extra-mitochondrial functions, such as involvement in apoptosis, protein biosynthesis and signal transduction. Dysregulations of the MRPs are associated with severe pathological conditions, including cancer. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

ABSTRACT MITCHELL III, ROBERT DRAKE. Global Human Health
ABSTRACT MITCHELL III, ROBERT DRAKE. Global Human Health Risks for Arthropod Repellents or Insecticides and Alternative Control Strategies. (Under the direction of Dr. R. Michael Roe). Protein-coding genes and environmental chemicals. New paradigms for human health risk assessment of environmental chemicals emphasize the use of molecular methods and human-derived cell lines. In this study, we examined the effects of the insect repellent DEET (N, N-diethyl-m-toluamide) and the phenylpyrazole insecticide fipronil (fluocyanobenpyrazole) on transcript levels in primary human hepatocytes. These chemicals were tested individually and as a mixture. RNA-Seq showed that 100 µM DEET significantly increased transcript levels for 108 genes and lowered transcript levels for 64 genes and fipronil at 10 µM increased the levels of 2,246 transcripts and decreased the levels for 1,428 transcripts. Fipronil was 21-times more effective than DEET in eliciting changes, even though the treatment concentration was 10-fold lower for fipronil versus DEET. The mixture of DEET and fipronil produced a more than additive effect (levels increased for 3,017 transcripts and decreased for 2,087 transcripts). The transcripts affected in our treatments influenced various biological pathways and processes important to normal cellular functions. Long non-protein coding RNAs and environmental chemicals. While the synthesis and use of new chemical compounds is at an all-time high, the study of their potential impact on human health is quickly falling behind. We chose to examine the effects of two common environmental chemicals, the insect repellent DEET and the insecticide fipronil, on transcript levels of long non-protein coding RNAs (lncRNAs) in primary human hepatocytes. -

Genome-Wide Analysis of Transcriptional Bursting-Induced Noise in Mammalian Cells
bioRxiv preprint doi: https://doi.org/10.1101/736207; this version posted August 15, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Title: Genome-wide analysis of transcriptional bursting-induced noise in mammalian cells Authors: Hiroshi Ochiai1*, Tetsutaro Hayashi2, Mana Umeda2, Mika Yoshimura2, Akihito Harada3, Yukiko Shimizu4, Kenta Nakano4, Noriko Saitoh5, Hiroshi Kimura6, Zhe Liu7, Takashi Yamamoto1, Tadashi Okamura4,8, Yasuyuki Ohkawa3, Itoshi Nikaido2,9* Affiliations: 1Graduate School of Integrated Sciences for Life, Hiroshima University, Higashi-Hiroshima, Hiroshima, 739-0046, Japan 2Laboratory for Bioinformatics Research, RIKEN BDR, Wako, Saitama, 351-0198, Japan 3Division of Transcriptomics, Medical Institute of Bioregulation, Kyushu University, Fukuoka, Fukuoka, 812-0054, Japan 4Department of Animal Medicine, National Center for Global Health and Medicine (NCGM), Tokyo, 812-0054, Japan 5Division of Cancer Biology, The Cancer Institute of JFCR, Tokyo, 135-8550, Japan 6Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Yokohama, Kanagawa, 226-8503, Japan 7Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, 20147, USA 8Section of Animal Models, Department of Infectious Diseases, National Center for Global Health and Medicine (NCGM), Tokyo, 812-0054, Japan 9Bioinformatics Course, Master’s/Doctoral Program in Life Science Innovation (T-LSI), School of Integrative and Global Majors (SIGMA), University of Tsukuba, Wako, 351-0198, Japan *Corresponding authors Corresponding authors e-mail addresses Hiroshi Ochiai, [email protected] Itoshi Nikaido, [email protected] bioRxiv preprint doi: https://doi.org/10.1101/736207; this version posted August 15, 2019. -

Browsing Genes and Genomes with Ensembl
The Bioinformatics Roadshow Tórshavn, The Faroe Islands 28-29 November 2012 BROWSING GENES AND GENOMES WITH ENSEMBL EXERCISES AND ANSWERS 1 BROWSER 3 BIOMART 8 VARIATION 13 COMPARATIVE GENOMICS 18 2 Note: These exercises are based on Ensembl version 69 (October 2012). After in future a new version has gone live, version 69 will still be available at http://e69.ensembl.org/. If your answer doesn’t correspond with the given answer, please consult the instructor. ______________________________________________________________ BROWSER ______________________________________________________________ Exercise 1 – Exploring a gene (a) Find the human F9 (coagulation factor IX) gene. On which chromosome and which strand of the genome is this gene located? How many transcripts (splice variants) have been annotated for it? (b) What is the longest transcript? How long is the protein it encodes? Has this transcript been annotated automatically (by Ensembl) or manually (by Havana)? How many exons does it have? Are any of the exons completely or partially untranslated? (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. the N-terminal or C-terminal half) of the protein encoded by ENST00000218099 does its peptidase activity reside? (f) Have any missense variants been discovered for the protein encoded by ENST00000218099? (g) Is there a mouse orthologue predicted for the human F9 gene? (h) If you have yourself a gene of interest, explore what information Ensembl displays about it! ______________________________________________________________ Answer (a) 8 Go to the Ensembl homepage (http://www.ensembl.org/). -
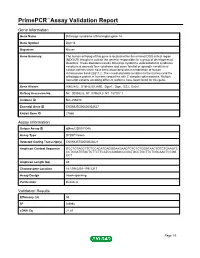
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DiGeorge syndrome critical region gene 14 Gene Symbol Dgcr14 Organism Mouse Gene Summary The human ortholog of this gene is located within the minimal DGS critical region (MDGCR) thought to contain the gene(s) responsible for a group of developmental disorders. These disorders include DiGeorge syndrome velocardiofacial syndrome conotruncal anomaly face syndrome and some familial or sporadic conotruncal cardiac defects which have been associated with microdeletion of human chromosome band 22q11.2. The encoded protein localizes to the nucleus and the orthologous protein in humans co-purifies with C complex spliceosomes. Multiple transcript variants encoding different isoforms have been found for this gene. Gene Aliases AI462402, D16H22S1269E, Dgcr1, Dgsi, ES2, Es2el RefSeq Accession No. NC_000082.6, NT_039624.8, NT_187007.1 UniGene ID Mm.256480 Ensembl Gene ID ENSMUSG00000003527 Entrez Gene ID 27886 Assay Information Unique Assay ID qMmuCID0011048 Assay Type SYBR® Green Detected Coding Transcript(s) ENSMUST00000003621 Amplicon Context Sequence GCCTGTAGCTTCTCCACATCAGGGAAGAAGTCTCTCTGGATAACTGTCTGAAGTC CCTCGATGTACTCTTCTTCATCCAGGACCCGCTGCCTGCTTCTCGCAACTCCGG CCT Amplicon Length (bp) 82 Chromosome Location 16:17910201-17911217 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 95 R2 0.9994 cDNA Cq 21.87 Page 1/5 PrimePCR™Assay Validation Report cDNA Tm (Celsius) 79.5 gDNA Cq 23.94 Specificity (%) 100 Information to assist with data interpretation is provided -

Somatic Mutational Landscape of AML with Inv(16) Or T(8;21) Identifies Patterns of Clonal Evolution in Relapse Leukemia
Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Sood, R., N. F. Hansen, F. X. Donovan, B. Carrington, D. Bucci, B. Maskeri, A. Young, et al. 2015. “Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia.” Leukemia 30 (2): 501-504. doi:10.1038/ leu.2015.141. http://dx.doi.org/10.1038/leu.2015.141. Published Version doi:10.1038/leu.2015.141 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:27320188 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Leukemia Manuscript Author . Author manuscript; Manuscript Author available in PMC 2016 May 18. Published in final edited form as: Leukemia. 2016 February ; 30(2): 501–504. doi:10.1038/leu.2015.141. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia Raman Sood1,4, Nancy F. Hansen2, Frank X. Donovan3, Blake Carrington4, Donna Bucci5, Baishali Maskeri6, Alice Young6, Niraj S. Trivedi7, Jessica Kohlschmidt5,8, Richard M. Stone9, Michael A. Caligiuri5, Settara C. Chandrasekharappa3,10, Guido Marcucci5,11, James C. -

CRNKL1 Is a Highly Selective Regulator of Intron-Retaining HIV-1 and Cellular Mrnas
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 CRNKL1 is a highly selective regulator of intron-retaining HIV-1 and cellular mRNAs 2 3 4 Han Xiao1, Emanuel Wyler2#, Miha Milek2#, Bastian Grewe3, Philipp Kirchner4, Arif Ekici4, Ana Beatriz 5 Oliveira Villela Silva1, Doris Jungnickl1, Markus Landthaler2,5, Armin Ensser1, and Klaus Überla1* 6 7 1 Institute of Clinical and Molecular Virology, University Hospital Erlangen, Friedrich-Alexander 8 Universität Erlangen-Nürnberg, Erlangen, Germany 9 2 Berlin Institute for Medical Systems Biology, Max-Delbrück-Center for Molecular Medicine in the 10 Helmholtz Association, Robert-Rössle-Strasse 10, 13125, Berlin, Germany 11 3 Department of Molecular and Medical Virology, Ruhr-University, Bochum, Germany 12 4 Institute of Human Genetics, University Hospital Erlangen, Friedrich-Alexander Universität Erlangen- 13 Nürnberg, Erlangen, Germany 14 5 IRI Life Sciences, Institute für Biologie, Humboldt Universität zu Berlin, Philippstraße 13, 10115, Berlin, 15 Germany 16 # these two authors contributed equally 17 18 19 *Corresponding author: 20 Prof. Dr. Klaus Überla 21 Institute of Clinical and Molecular Virology, University Hospital Erlangen 22 Friedrich-Alexander Universität Erlangen-Nürnberg 23 Schlossgarten 4, 91054 Erlangen 24 Germany 25 Tel: (+49) 9131-8523563 26 e-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.02.04.934927; this version posted February 11, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. -

Emerging Roles of Metallothioneins in Beta Cell Pathophysiology: Beyond and Above Metal Homeostasis and Antioxidant Response
biology Review Emerging Roles of Metallothioneins in Beta Cell Pathophysiology: Beyond and above Metal Homeostasis and Antioxidant Response Mohammed Bensellam 1,* , D. Ross Laybutt 2,3 and Jean-Christophe Jonas 1 1 Pôle D’endocrinologie, Diabète et Nutrition, Institut de Recherche Expérimentale et Clinique, Université Catholique de Louvain, B-1200 Brussels, Belgium; [email protected] 2 Garvan Institute of Medical Research, Sydney, NSW 2010, Australia; [email protected] 3 St Vincent’s Clinical School, UNSW Sydney, Sydney, NSW 2010, Australia * Correspondence: [email protected]; Tel.: +32-2764-9586 Simple Summary: Defective insulin secretion by pancreatic beta cells is key for the development of type 2 diabetes but the precise mechanisms involved are poorly understood. Metallothioneins are metal binding proteins whose precise biological roles have not been fully characterized. Available evidence indicated that Metallothioneins are protective cellular effectors involved in heavy metal detoxification, metal ion homeostasis and antioxidant defense. This concept has however been challenged by emerging evidence in different medical research fields revealing novel negative roles of Metallothioneins, including in the context of diabetes. In this review, we gather and analyze the available knowledge regarding the complex roles of Metallothioneins in pancreatic beta cell biology and insulin secretion. We comprehensively analyze the evidence showing positive effects Citation: Bensellam, M.; Laybutt, of Metallothioneins on beta cell function and survival as well as the emerging evidence revealing D.R.; Jonas, J.-C. Emerging Roles of negative effects and discuss the possible underlying mechanisms. We expose in parallel findings Metallothioneins in Beta Cell from other medical research fields and underscore unsettled questions. -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas.