Decision Report
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Volume 5 Pt 3
Conservation Science W. Aust. 7 (3) : 583–592 (2010) Flora and vegetation of banded iron formations of the Yilgarn Craton: Mt Ida Greenstone Belt and Mt Hope RACHEL MEISSNER AND GAYNOR OWEN Science Division, Department of Environment and Conservation, PO Box 51, Wanneroo, Western Australia, 6946. Email: [email protected] ABSTRACT The Mount Ida greenstone belt is located within the Southern Cross Granite-Greenstone terrane (Wyche 2003) and is expressed as low ranges of banded ironstone formation. Recent increases in the value of iron ore have made the mining of banded iron formations more attractive. This paper describes the flora and vegetation on these ranges. A total eighty seven taxa, including one annual, were recorded from the ranges. One priority species (Atkins 2008), Calytrix erosipetala, and two range extensions were recorded. Hierarchical classification identified four plant communities on the ranges. The dominant floristic communities occurred on the slopes and crests (Community 2) and on the lower and colluvial slopes (Community 3). Upland communities were less fertile and there was a lower concentration of soil mobile elements than in the lowland communities. Currently, the Mt Ida Greenstone Belt is not within a conservation estate. Keywords: BIF, banded ironstone, floristic communities, ranges, Yilgarn INTRODUCTION The main range of the Mt Ida Greenstone Belt can be divided into two areas; the Mount Mason range in the Banded iron formations (BIF) are ancient sedimentary north extending 13km, and the Mt Ida Range, south of rocks formed in the Archaean period approximately 3.8 Mt Mason and extending c. 7km. The Mt Mason range is to 2.5 billion years ago. -

Charles Darwin Reserve
CHARLES DARWIN RESERVE (WHITE WELLS STATION) WESTERN AUSTRALIA FIELD HERBARIUM Volunteers of the Bushland Plant Survey Project Wildflower Society of Western Australia (Inc.) PO Box 519 Floreat WA 6014 for Bush Heritage Australia July 2010 This project was supported by the Wildflower Society of Western Australia Support was also provided by the WA Department of Environment and Conservation NOTE: This Field Herbarium is to remain the property of Bush Heritage, in so long as the Reserve is managed sympathetically with the bushland, and the owners are able to care for the Herbarium so it does not deteriorate. In the event these criteria cannot be met the Field Herbarium is to be handed over to the Geraldton Regional Herbarium. For further information contact the WA Herbarium, Department of Environment and Conservation, Locked Bag 104, Bentley Delivery Centre, WA 6983 Phone (08) 9334 0500. Charles Darwin Reserve (White Wells Station), Western Australia – Field Herbarium CONTENTS 1 BACKGROUND AND ACKNOWLEDGEMENTS..................................................................................... 1 Map 1 Wildflower Society of WA survey sites at Charles Darwin Reserve - August 2008 .......................... 2 Map 2 Wildflower Society of WA survey sites at Charles Darwin Reserve – October 2008 ........................ 3 2 FLORA ........................................................................................................................................................... 4 3 THE FIELD HERBARIUM .......................................................................................................................... -

Geraldton Mt Magnet Road SLK 222.45 Biological Survey May 2012
Main Roads Western Australia Report for Material Source Area: Geraldton Mt Magnet Road SLK 222.45 Biological Survey May 2012 This Report: has been prepared by GHD for Main Roads Western Australia (MRWA) and may only be used and relied on by MRWA for the purpose agreed between GHD and MRWA as set out in section 1.3 of this Report. GHD otherwise disclaims responsibility to any person other than MRWA arising in connection with this Report. GHD also excludes implied warranties and conditions, to the extent legally permissible. The services undertaken by GHD in connection with preparing this Report were limited to those specifically detailed in the Report and are subject to the scope limitations set out in the Report. The opinions, conclusions and any recommendations in this Report are based on conditions encountered and information reviewed at the date of preparation of the Report. GHD has no responsibility or obligation to update this Report to account for events or changes occurring subsequent to the date that the Report was prepared. The opinions, conclusions and any recommendations in this Report are based on assumptions made by GHD described in this Report. GHD disclaims liability arising from any of the assumptions being incorrect. GHD has prepared this Report on the basis of information provided by MRWA and others who provided information to GHD (including Government authorities), which GHD has not independently verified or checked beyond the agreed scope of work. GHD does not accept liability in connection with such unverified information, including errors and omissions in the Report which were caused by errors or omissions in that information.” The opinions, conclusions and any recommendations in this Report are based on information obtained from, and testing undertaken at or in connection with, specific sample points. -

Clearing Permit Decision Report
Clearing Permit Decision Report 1. Application details 1.1. Permit application details Permit application No.: 8786/1 Permit type: Purpose Permit 1.2. Proponent details Proponent’s name: Mid-West Tungsten Pty Ltd 1.3. Property details Property: Mining Lease 59/386 Mining Lease 59/387 Mining Lease 59/425 Miscellaneous Licence 59/161 Miscellaneous Licence 59/162 Local Government Area: Shire of Perenjori Colloquial name: Mt Mulgine Project 1.4. Application Clearing Area (ha) No. Trees Method of Clearing For the purpose of: 201.8 Mechanical Removal Mineral Production and Associated Activities 1.5. Decision on application Decision on Permit Application: Refuse Decision Date: 24 September 2020 Reasons for Decision The clearing permit application was received on 08 January 2020 and has been assessed against the clearing principles, planning instruments, and other matters in accordance with section 51O of the Environmental Protection Act 1986 (EP Act). The initial assessment of the application determined that the proposed clearing would likely result in significant impacts to conservation significant flora and fauna, and was therefore unlikely to be considered acceptable. The applicant was given the opportunity to provide additional information and/or modify their clearing proposal in order to reduce the environmental impacts. In response, Mid-West Tungsten Pty Ltd provided additional information and biological survey reports. The additional information and survey reports were taken into consideration when making the decision on this application. -

Employing Hypothesis Testing and Data from Multiple Genomic Compartments to Resolve Recalcitrant Backbone Nodes in Goodenia S.L. T (Goodeniaceae) ⁎ Rachel S
Molecular Phylogenetics and Evolution 127 (2018) 502–512 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Employing hypothesis testing and data from multiple genomic compartments to resolve recalcitrant backbone nodes in Goodenia s.l. T (Goodeniaceae) ⁎ Rachel S. Jabailya,b, , Kelly A. Shepherdc, Pryce S. Michenerb, Caroline J. Bushb, Rodrigo Riverod,e, Andrew G. Gardnerf, Emily B. Sessad,g a Department of Organismal Biology & Ecology, Colorado College, Colorado Springs, CO 80903, USA b Department of Biology, Rhodes College, Memphis, TN 38112, USA c Western Australian Herbarium, Department of Biodiversity, Conservation and Attractions, Kensington, WA 6151, Australia d Department of Biology, University of Florida, Gainesville, FL 32607, USA e Department of Natural Resources and Environmental Management, University of Hawaii– Mānoa, Honolulu, HI 96822, USA f Department of Biological Sciences, California State University, Stanislaus, One University Circle, Turlock, CA 95382, USA g Genetics Institute, University of Florida, Gainesville, FL 32607, USA ARTICLE INFO ABSTRACT Keywords: Goodeniaceae is a primarily Australian flowering plant family with a complex taxonomy and evolutionary Chloroplast genome history. Previous phylogenetic analyses have successfully resolved the backbone topology of the largest clade in Conserved ortholog set (COS) the family, Goodenia s.l., but have failed to clarify relationships within the species-rich and enigmatic Goodenia Genome skimming clade C, a prerequisite for taxonomic revision of the group. We used genome skimming to retrieve sequences for Goodeniaceae chloroplast, mitochondrial, and nuclear markers for 24 taxa representing Goodenia s.l., with a particular focus on Mitochondrial genome Goodenia clade C. We performed extensive hypothesis tests to explore incongruence in clade C and evaluate Phylogeny Rapid radiation statistical support for clades within this group, using datasets from all three genomic compartments. -

Charles Darwin, Kadji Kadji, Karara, Lochada Reserves WA
BUSH BLITZ SPECIES DISCOVERY PROGRAM Charles Darwin Reserve WA 3–9 May · 14–25 September · 7–18 December 2009 Kadji Kadji, Karara, Lochada Reserves WA 14–25 September · 7–18 December 2009 What is Contents Bush Blitz? Bush Blitz is a four-year, What is Bush Blitz 2 multi-million dollar Summary 3 partnership between the Abbreviations 3 Australian Government, Introduction 4 BHP Billiton, and Earthwatch Reserves Overview 5 Australia to document plants Methods 8 and animals in selected properties across Australia’s Results 10 National Reserve System. Discussion 12 Appendix A: Species Lists 15 Fauna 16 This innovative partnership Vertebrates 16 harnesses the expertise of many Invertebrates 25 of Australia’s top scientists from Flora 48 museums, herbaria, universities, Appendix B: Rare and Threatened Species 79 and other institutions and Fauna 80 organisations across the country. Flora 81 Appendix C: Exotic and Pest Species 83 Fauna 84 Flora 85 2 Bush Blitz survey report Summary Bush Blitz fieldwork was conducted at four National Reserve System properties in the Western Australian Avon Wheatbelt and Yalgoo Bioregions during 2009. This included a pilot study Abbreviations at Charles Darwin Reserve and a longer study of Charles Darwin, Kadji Kadji, Lochada and Karara reserves. Results include 651 species added to those known across the reserves and the discovery of 35 putative species new to science. The majority of ANHAT these new species occur within the heteroptera (plant bugs) and Australian Natural Heritage Assessment lepidoptera (butterflies and moths) taxonomic groups. Tool Malleefowl (Leipoa ocellata), listed as vulnerable under the EPBC Act federal Environmental Protection and Biodiversity Conservation Environment Protection and Biodiversity Act 1999 (EPBC Act), were observed on Charles Darwin Reserve. -
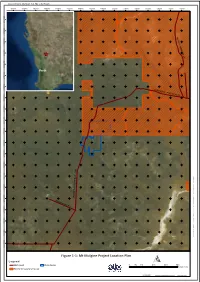
Tungsten Mining NL Mt Mulgine Project EPA REFERRAL
Document Name: 20170125_TUN_Fig1-1_Biol Report 482000 484000 486000 488000 490000 492000 494000 496000 498000 500000 502000 504000 506000 508000 510000 512000 0 0 0 2 9 7 6 0 0 0 0 9 7 6 Y A 0 L 0 G 0 O 8 8 O 7 6 - N I N G 0 0 H 0 A 6 N 8 7 R 6 ^_ D 0 0 0 4 8 7 6 Perth 0 0 0 2 8 7 6 0 0 0 0 8 7 6 W ARRIE DAR - PERE 0 NJO 0 RI RD 0 8 7 7 6 0 0 0 6 7 7 6 0 0 0 4 7 7 6 0 0 0 2 7 7 6 0 0 0 0 7 7 6 0 0 0 8 6 7 6 0 0 0 6 6 7 6 D R E 0 0 N I \ 0 \ M 4 M A 6 I N 7 R S 6 E E R V P E R P - P C O \ 0 s e C 0 r v 0 e R r 2 s t 6 A o r 7 a D g 6 e E \ I A P R M R G I A S 0 a 0 n W d 0 M 0 a 6 p 7 p i n 6 B O g O \ N 0 E 3 R _ O C N l i G e n 0 R D t \ 0 T U 0 N 8 \ 0 5 2 7 _ G 6 I S M a p s \ 2 0 0 1 0 7 0 0 1 6 2 5 5 _ T 7 U 6 N _ F i g 1 - 1 _ B 0 i o 0 l R 0 e 4 p o 5 r t 7 . -

The Unexpected Depths of Genome-Skimming Data: a Case Study Examining Goodeniaceae Floral Symmetry Genes Author(S): Brent A
The Unexpected Depths of Genome-Skimming Data: A Case Study Examining Goodeniaceae Floral Symmetry Genes Author(s): Brent A. Berger, Jiahong Han, Emily B. Sessa, Andrew G. Gardner, Kelly A. Shepherd, Vincent A. Ricigliano, Rachel S. Jabaily, and Dianella G. Howarth Source: Applications in Plant Sciences, 5(10) Published By: Botanical Society of America https://doi.org/10.3732/apps.1700042 URL: http://www.bioone.org/doi/full/10.3732/apps.1700042 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Applications in Plant Sciences 2017 5(10): 1700042 Applications in Plant Sciences APPLICATION ARTICLE THE UNEXPECTED DEPTHS OF GENOME-SKIMMING DATA: A CASE 1 STUDY EXAMINING GOODENIACEAE FLORAL SYMMETRY GENES BRENT A. BERGER2,8, JIAHONG HAN2, EMILY B. SEssa3, ANDREW G. GARDNER4, KELLY A. SHEPHERD5, VINCENT A. RICIGLIANO6, RACHEL S. JabaILY7, AND DIANELLA G. HOWARTH2 2Department of Biological Sciences, St. -

Rangelands, Western Australia
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

Declared Rare and Poorly Known Flora in the Geraldton District
WESTERN AUSTRALIAN WILDLIFE MANAGEMENT PROGRAM NO. 26 Declared Rare and Poorly Known Flora in the Geraldton District by Susan J. Patrick 2001 Department of Conservation and Land Management Locked Bag 104, Bentley Delivery Centre WA 6983 1 Department of Conservation and Land Management Locked Bag 104, Bentley Delivery Centre WA 6983 Department of Conservation and Land Management, Western Australia 2001 ISSN 0816-9713 Cover illustration: Verticordia spicata subsp. squamosa by Margaret Pieroni Editors ..........................................................................................................Angie Walker and Jill Pryde Page preparation ..................................................................................................................Angie Walker Maps ..................................................................................................... CALM Land Information Branch 2 FOREWORD Western Australian Wildlife Management Programs are a series of publications produced by the Department of Conservation and Land Management (CALM). The programs are prepared in addition to Regional Management Plans to provide detailed information and guidance for the management and protection of certain exploited or threatened species (e.g. Kangaroos, Noisy Scrub-bird and the Rose Mallee). This program provides a brief description of the appearance, distribution, habitat and conservation status of flora declared as rare under the Western Australian Wildlife Conservation Act (Threatened Flora) and poorly known flora (Priority -

Genetic Analyses of Casuarinas Using ISSR and FISSR Markers
Genetica 122: 161–172, 2004. 161 Ó 2004 Kluwer Academic Publishers. Printed in the Netherlands. Genetic analyses of Casuarinas using ISSR and FISSR markers R. Yasodha1, M. Kathirvel2, R. Sumathi1, K. Gurumurthi1, Sunil Archak2 & J. Nagaraju2,* 1Division of Plant Biotechnology, Institute of Forest Genetics and Tree Breeding, Coimbatore 641002, India; 2Laboratory of Molecular Genetics, Centre for DNA Fingerprinting and Diagnostics, Hyderabad 500076, India; *Author for correspondence (Phone: +91-40-715-1344; Fax: +91-40-715-5610; E-mails: jnagaraju@ cdfd.org.in, [email protected]) Received 11 August 2003 Accepted 2 March 2004 Key words: Allocasuarina, Casuarina, FISSR-PCR, ISSR-PCR Abstract Inter simple sequence repeat polymerase chain reaction (ISSR-PCR) was used for the genetic analysis of the six species of Allocasuarina, five species of Casuarina and 12 superior performing selections of C. equis- etifolia L. We also fingerprinted C. equisetifolia L. selections using Fluorescent-ISSR-PCR (FISSR-PCR), an improvised ISSR-PCR assay. The ISSR analysis provided information on the frequency of various simple sequence repeats in the casuarina genome. The di-nucleotide repeats were more common, among which (CA)n and its complementary nucleotide (GT)n repeat motifs amplified relatively higher number of bands with an average of 6.0 ± 3.5 and 6.3 ± 1.8 respectively. Eleven species of casuarinas were amplified with 10 primers anchored either at 5¢ or 3¢ end. A total of 253 PCR products were obtained and all were polymorphic, out of which 48 were specific to Allocasuarina and 36 were specific to Casuarina genus. Genetic similarity among the species was 0.251. -

Species List
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations.