Exoplanet Biosignatures: Future Directions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Life Beyond Earth the Search for Habitable Worlds in the Universe
C:/ITOOLS/WMS/CUP-NEW/4181325/WORKINGFOLDER/COEN/9781107026179HTL.3D i [1–2] 27.6.2013 10:59AM Life Beyond Earth The Search for Habitable Worlds in the Universe With current missions to Mars and the Earth-like moon Titan, and many more missions planned, humankind stands on the verge of exciting progress and possible major discoveries in our quest for life in space. What is life and where can it exist? What searches are being made to identify conditions for life on other worlds? If extraterrestrial inhabited worlds are found, how can we explore them? Could humans survive beyond the Earth? In this book, two leading astrophysicists provide an engaging account of where we stand in our quest for habitable environments, in the Solar System and beyond. Starting from basic concepts, the narrative builds up scientifically, including more in-depth material as boxed additions to the main text. The authors recount fascinating recent discoveries, arising from space missions and from observations using ground-based telescopes, of possible life-related artefacts in Martian meteorites, of extrasolar planets, and of subsurface oceans on Europa, Titan and Enceladus. They also provide a forward look to exciting future missions, including the return to Venus, Mars and the Moon; further explorations of Pluto and Jupiter’s icy moons; and placing giant planet-seeking telescopes in orbit beyond Jupiter. showing how we approach the question of finding out whether the life that teems on our own planet is unique. This is an exciting, informative read for anyone interested in the search for habitable and inhabited planets, and makes an excellent primer for students keen to learn about astrobiology, habitability, planetary science and astronomy. -

Distribution of Nitrifying Bacteria Under Fluctuating Environmental Conditions
Distribution of Nitrifying Bacteria Under Fluctuating Environmental Conditions Joseph M. Battistelli Stratford, Connecticut B.S., Rutgers, the State Universi ty of New Jersey, New Brunswick Campus, 2002 A Dissertation presented to the Graduate Faculty of the University of Virginia in Candidacy for the Degree of Doctor of Philosophy Department of Environmental Sciences University of Virginia December, 2012 ii Abstract Engineered systems that mimic tidal wetlands are ideal for point-of-source treatment of wastewater due to their low energy requirements and small physical footprint. In this study, vertical columns were constructed to mimic the flood-and-drain cycles of tidal wetland treatment systems (TWTS) in order to study how variations in the frequency and duration of flooding affect the efficiency of microbially-mediated nitrogen + removal from synthetic wastewater (containing whey protein and NH 4 ). Altering the frequency of flooding, which determines the temporal juxtaposition of aerobic and anaerobic conditions in the reactor, had a significant effect on overall nitrogen removal; + columns more frequent cycling were very efficient at converting the NH4 in the feed to - - NO 3 . At a flooding frequency of 8-cycles per day, NO 3 began to disappear from the systems in both High- and Low - N treatments. The longer flooding duration appeared to increased anaerobiosis and allowed denitrification to proceed more effectively, while allowing nitrification to proceed when oxygen was available. Analysis of depth profiles of abundance revealed distinct differences between the tidal and trickling systems abundance profiles. Overall, these results demonstrate a tight coupling of environmental conditions with the abundance of ammonium oxidizing bacteria and suggest several experimental modifications, such variable tidal cycles, could be implemented to enhance the functioning of TWTS. -

Astrobiologia E a Busca Científica De Vida Fora Da Terra
Astronomia ao Meio Dia – IAG-USP 05/10/2017 Astrobiologia e a busca científica de vida fora da Terra Fabio Rodrigues Instituto de Química - USP Núcleo de Pesquisa em Astrobiologia - USP [email protected] Vida Extraterrestre • Prêmio Guzman (1900): 100.000 francos oferecido pela Académie des sciences (França) para quem conseguisse estabelecer comunicação extraterrestre. Curiosidade: Em seu texto, o prêmio excluia explícitamente a comunicação com Marte, pois neste planeta era óbvio que havia vida!! Evolução das observações de Marte! Christiaan Huygens, 1659 Telescopic view, 1960s Giovanni Schiaparelli, 1888 Hubble Space Telescope, 1997 Mars Global Surveyor, 2002 Furo com 1,6 cm de diâmetro! Especulativa Experimental / observacional Foto obtida pelo Mars Science Laboratoy (Curiosity) em 12 de Maio de 2014! O que é astrobiologia? Não é uma nova disciplina, mas um novo enfoque a antigos temas Origem e evolução da vida Distribição da vida, na Terra e fora dela Futuro da vida O que é astrobiologia? Não é uma nova disciplina, mas um novo enfoque a antigos temas Astrobiólogo?? - Físicos, químicos, biólogos, astrônomos, geólogos, cientistas planetários, filósofos, etc. - Interessados nos problemas e com o enfoque proposto pela astrobiologia! Big Bang Estrelas e supernovas Formação dos elementos Big Bang Estrelas e supernovas Moléculas Formação dos elementos (Astroquímica) + + - + + + 2 átomos: AlO, C2, CH, CH , CN, CN , CN , CO, CO , CS, FeO, H2, HCl, HCl , HO, OH , KCl, NH, + + N2, NO, NO , NaCl, MgH , O2, PO, SH, SO, SiC, SiN, SiO, TiO etc. + + 3 átomos: AlOH, C3, C2H, C2O, C2P, CO2, H3 , H2C, H2O, H2O , HO2, H2S, HCN, HNC, HCO, + + HCO+, HCP, HNC, HN2 , MgCN, NH2, N2H , N2O, NaOH, O3, SO2, SiC2, SiCN, TiO2 etc. -

Development of Electrochemical Methods for Detection of Pesticides and Biofuel
Development of Electrochemical Methods for Detection of Pesticides and Biofuel Production Asli Sahin Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2012 © 2012 Asli Sahin All Rights Reserved ABSTRACT Development of Electrochemical Methods for Detection of Pesticides and Biofuel Production Asli Sahin Electrochemical methods coupled with biological elements are used in industrial, medical and environmental applications. In this thesis we will discuss two such applications: an electrochemical biosensor for detection of pesticides and biofuel generation using electrochemical methods coupled with microorganisms. Electrochemical biosensors are commonly used as a result of their selectivity, sensitivity, rapid response and portability. A common application for electrochemical biosensors is detection of pesticides and toxins in water samples. In this thesis, we will focus on detection of organosphosphates (OPs), a group of compounds that are commonly used as pesticides and nerve agents. Rapid and sensitive detection of these compounds has been an area of active research due to their high toxicity. Amperometric and potentiometric electrochemical biosensors that use organophosphorus hydrolase (OPH), an enzyme that can hydrolyze a broad class of OPs, have been reported for field detection of OPs. Amperometry is used for detection of electroactive leaving groups and potentiometry is used for detection of pH changes that take place during the hydrolysis reaction. Both these methods have limitations: using amperometric biosensors, very low limits of detection are achieved but this method is limited to the few OPs with electroactive leaving groups, on the other hand potentiometric biosensor can be used for detection of all OPs but they don’t have low enough limits of detection. -

Outline of Physical Science
Outline of physical science “Physical Science” redirects here. It is not to be confused • Astronomy – study of celestial objects (such as stars, with Physics. galaxies, planets, moons, asteroids, comets and neb- ulae), the physics, chemistry, and evolution of such Physical science is a branch of natural science that stud- objects, and phenomena that originate outside the atmosphere of Earth, including supernovae explo- ies non-living systems, in contrast to life science. It in turn has many branches, each referred to as a “physical sions, gamma ray bursts, and cosmic microwave background radiation. science”, together called the “physical sciences”. How- ever, the term “physical” creates an unintended, some- • Branches of astronomy what arbitrary distinction, since many branches of physi- cal science also study biological phenomena and branches • Chemistry – studies the composition, structure, of chemistry such as organic chemistry. properties and change of matter.[8][9] In this realm, chemistry deals with such topics as the properties of individual atoms, the manner in which atoms form 1 What is physical science? chemical bonds in the formation of compounds, the interactions of substances through intermolecular forces to give matter its general properties, and the Physical science can be described as all of the following: interactions between substances through chemical reactions to form different substances. • A branch of science (a systematic enterprise that builds and organizes knowledge in the form of • Branches of chemistry testable explanations and predictions about the • universe).[1][2][3] Earth science – all-embracing term referring to the fields of science dealing with planet Earth. Earth • A branch of natural science – natural science science is the study of how the natural environ- is a major branch of science that tries to ex- ment (ecosphere or Earth system) works and how it plain and predict nature’s phenomena, based evolved to its current state. -

Vida En Otros Planetas Eduardo Battaner Universidad De Granada
Vida en otros planetas Eduardo Battaner Universidad de Granada Estudiantes de Física y Electrónica EFE. Vida. Granada, 2011 ¿Vida en otros planetas? ● Sólo tenemos una prueba observacional. ● No sabemos cómo se produce la vida. Muchas gracias EFE. Vida. Granada, 2011 Hay planetas extrasolares ● Se han descubierto unos 500. ● Técnicas: Observación directa, efecto Doppler, astrometría, tránsitos. ● Corot, Kepler, Darwin (>2014) (espectrometría de atmósferas de planetas terrestres). ● Pero tienen vida? EFE. Vida. Granada, 2011 Misión Darwin ● ESA, 2014 ● 3 telescopios de 3m en interferometría, mili arcsec. ● Situado en L2 ● Infrarrojo. La estrella brilla sólo 1 millón de veces más. ● La estrella se anula interferométricamente. ● Espectro IR, oxígeno? Agua? ● La fotosíntesis produce oxígeno en La Tierra. ● Aunque puede producirse por fotodisociación del CO2. EFE. Vida. Granada, 2011 EFE. Vida. Granada, 2011 Titan ● Descubierto por Huygens (1655) ● Tiene atmósfera, Comás Solá (1908), por medio de ocultación. ● Diámetro 2000 km. Temperatura 90K ● Atmósfera de nitrógeno (94%) y metano. ● Orbitador Cassini, Sonda Huygens (2005) ● Montañas, ríos y mares (metano), islas ● Guijarros de hielo EFE. Vida. Granada, 2011 Titán Hicrocarburos procedentes de disociación de CH4 ¿Cuál es la fuente de CH4? Vientos muy rápidos como en Venus Tormentas: Sánchez Lavega, con descargas bruscas de metano. El CH4 se filtra por el suelo. EFE. Vida. Granada, 2011 EFE. Vida. Granada, 2011 ● ¿Fórmula de Drake ? ● ¿ªZona de habitabilidadº? ● ¿Oparin y Haldane? ● ¿Experimento de Miller y Urey? ● ¿Panspermia? ● Paradoja de Fermi. ● La ausencia de evidencia ¿es la evidencia de la ausencia? EFE. Vida. Granada, 2011 Gliese 581g ● Catálogo de Wilhelm Gliese (1957) < 25 pc ● Gliese 581 es de tipo M2 (-31ë , -12ë), 37 días. -
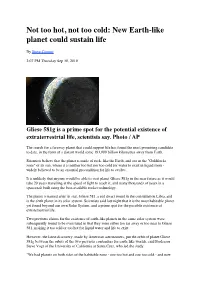
Goldilocks Planet
Not too hot, not too cold: New Earth-like planet could sustain life By Steve Connor 2:07 PM Thursday Sep 30, 2010 Gliese 581g is a prime spot for the potential existence of extraterrestrial life, scientists say. Photo / AP The search for a faraway planet that could support life has found the most promising candidate to date, in the form of a distant world some 193,000 billion kilometres away from Earth. Scientists believe that the planet is made of rock, like the Earth, and sits in the "Goldilocks zone" of its sun, where it is neither too hot nor too cold for water to exist in liquid form - widely believed to be an essential precondition for life to evolve. It is unlikely that anyone would be able to visit planet Gliese 581g in the near future as it would take 20 years travelling at the speed of light to reach it, and many thousands of years in a spacecraft built using the best-available rocket technology. The planet is named after its star, Gliese 581, a red dwarf found in the constellation Libra, and is the sixth planet in its solar system. Scientists said last night that it is the most habitable planet yet found beyond our own Solar System, and a prime spot for the possible existence of extraterrestrial life. Two previous claims for the existence of earth-like planets in the same solar system were subsequently found to be overstated in that they were either too far away or too near to Gliese 581, making it too cold or too hot for liquid water and life to exist. -

Metal Complexes of Penicillin and Cephalosporin Antibiotics
I METAL COMPLEXES OF PENICILLIN AND CEPHALOSPORIN ANTIBIOTICS A thesis submitted to THE UNIVERSITY OF CAPE TOWN in fulfilment of the requirement$ forTown the degree of DOCTOR OF PHILOSOPHY Cape of by GRAHAM E. JACKSON University Department of Chernis try, University of Cape Town, Rondebosch, Cape, · South Africa. September 1975. The copyright of th:s the~is is held by the University of C::i~r:: To\vn. Reproduction of i .. c whole or any part \ . may be made for study purposes only, and \; not for publication. The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgementTown of the source. The thesis is to be used for private study or non- commercial research purposes only. Cape Published by the University ofof Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University '· ii ACKNOWLEDGEMENTS I would like to express my sincere thanks to my supervisors: Dr. L.R. Nassimbeni, Dr. P.W. Linder and Dr. G.V. Fazakerley for their invaluable guidance and friendship throughout the course of this work. I would also like to thank my colleagues, Jill Russel, Melanie Wolf and Graham Mortimor for their many useful conrrnents. I am indebted to AE & CI for financial assistance during the course of. this study. iii ABSTRACT The interaction between metal"'.'ions and the penici l)in and cephalosporin antibiotics have been studied in an attempt to determine both the site and mechanism of this interaction. The solution conformation of the Cu(II) and Mn(II) complexes were determined using an n.m.r, line broadening, technique. -
![Full Text [PDF]](https://docslib.b-cdn.net/cover/9721/full-text-pdf-1179721.webp)
Full Text [PDF]
® The Americas Journal of Plant Science and Biotechnology ©2011 Global Science Books Living in the O-zone: Ozone Formation, Ozone-Plant Interactions and the Impact of Ozone Pollution on Plant Homeostasis Godfrey P. Miles1 • Marcus A. Samuel2* 1 U.S. Department of Agriculture – Agricultural Research Service, 5230 Konnowac Pass Road, Wapato, WA 98951, USA 2 Department of Biological Sciences, University of Calgary, 2500, University Drive, NW, Calgary, AB, T2N 1N4, Canada Corresponding author : * [email protected] ABSTRACT Ozone (O3) is a key constituent of the terrestrial atmosphere. Unlike in the stratosphere, where O3 provides an essential barrier to incoming UV radiation, within the troposphere, it is a major secondary air pollutant that is estimated to cause more damage to plant life than all other air pollutants combined. In the troposphere, O3 is produced by photochemical oxidation of primary precursor emissions of volatile organic compounds (VOCs), carbon monoxide (CO), and sulfur dioxide (SO2) in association with elevated levels of oxides of nitrogen (NOx NO + NO2). Because of its strong oxidizing potential, ozone is damaging to plant life through oxidative damage to proteins, nucleic acids and lipids either directly or as a result of reactive oxygen species (ROS) derived from O3 decomposition. In plants, ROS, directly or indirectly derived from O3 exposure, are routinely scavenged by an array of enzymatic and non-enzymatic antioxidant defense mechanisms. The various ROS generated by O3 have strong influence on the plant’s biochemical -

The Lick-Carnegie Exoplanet Survey: a 3.1 M Earth Planet in The
The Lick-Carnegie Exoplanet Survey: A 3.1 M⊕ Planet in the Habitable Zone of the Nearby M3V Star Gliese 581 Steven S. Vogt1, R. Paul Butler2, E. J. Rivera1, N. Haghighipour3, Gregory W. Henry4, and Michael H. Williamson4 Received ; accepted arXiv:1009.5733v1 [astro-ph.EP] 29 Sep 2010 1UCO/Lick Observatory, University of California, Santa Cruz, CA 95064 2Department of Terrestrial Magnetism, Carnegie Institution of Washington, 5241 Broad Branch Road, NW, Washington, DC 20015-1305 3Institute for Astronomy and NASA Astrobiology Institute, University of Hawaii-Manoa, Honolulu, HI 96822 4Tennessee State University, Center of Excellence in Information Systems, 3500 John A. Merritt Blvd., Box 9501, Nashville, TN. 37209-1561 –2– ABSTRACT We present 11 years of HIRES precision radial velocities (RV) of the nearby M3V star Gliese 581, combining our data set of 122 precision RVs with an ex- isting published 4.3-year set of 119 HARPS precision RVs. The velocity set now indicates 6 companions in Keplerian motion around this star. Differential photometry indicates a likely stellar rotation period of ∼ 94 days and reveals no significant periodic variability at any of the Keplerian periods, supporting planetary orbital motion as the cause of all the radial velocity variations. The combined data set strongly confirms the 5.37-day, 12.9-day, 3.15-day, and 67-day planets previously announced by Bonfils et al. (2005), Udry et al. (2007), and Mayor et al. (2009). The observations also indicate a 5th planet in the system, GJ 581f, a minimum-mass 7.0 M⊕ planet orbiting in a 0.758 AU orbit of period 433 days and a 6th planet, GJ 581g, a minimum-mass 3.1 M⊕ planet orbiting at 0.146 AU with a period of 36.6 days. -

Mathematics for Input Space Probes in the Atmosphere of Gliese 581D
Parana Journal of Science and Education, v.2, n.5, (6-13) July 14, 2016 PJSE, ISSN 2447-6153, c 2015-2016 https://sites.google.com/site/pjsciencea/ Mathematics for input space probes in the atmosphere of Gliese 581d R. Gobato1, A. Gobato2 and D. F. G. Fedrigo3 Abstract The work is a mathematical approach to the entry of an aerospace vehicle, as a probe or capsule in the atmosphere of the planet Gliese 581d, using data collected from the results of atmospheric models of the planet. GJ581d was the first planet candidate of a few Earth masses reported in the circum-stellar habitable zone of another star. It is located in the Gliese 581 star system, is a star red dwarf about 20 light years away from Earth in the constellation Libra. Its estimated mass is about a third of that of the Sun. It has been suggested that the recently discovered exoplanet GJ581d might be able to support liquid water due to its relatively low mass and orbital distance. However, GJ581d receives 35% less stellar energy than the planet Mars and is probably locked in tidal resonance, with extremely low insolation at the poles and possibly a permanent night side. The climate that demonstrate GJ581d will have a stable atmosphere and surface liquid water for a wide range of plausible cases, making it the first confirmed super-Earth (2-10 Earth masses) in the habitable zone. According to the general principle of relativity, “All systems of reference are equivalent with respect to the formulation of the fundamental laws of physics.” In this case all the equations studied apply to the exoplanet Gliese 581d. -

Open Research Online Oro.Open.Ac.Uk
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Open Research Online Open Research Online The Open University’s repository of research publications and other research outputs Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life Journal Item How to cite: Stevenson, Andrew; Burkhardt, Jürgen; Cockell, Charles S.; Cray, Jonathan A.; Dijksterhuis, Jan; Fox-Powell, Mark; Kee, Terence P.; Kminek, Gerhard; McGenity, Terry J.; Timmis, Kenneth N.; Timson, David J.; Voytek, Mary A.; Westall, Frances; Yakimov, Michail M. and Hallsworth, John E. (2015). Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life. Environmental Microbiology, 17(2) pp. 257–277. For guidance on citations see FAQs. c 2014 Society for Applied Microbiology and John Wiley Sons Ltd Version: Version of Record Link(s) to article on publisher’s website: http://dx.doi.org/doi:10.1111/1462-2920.12598 Copyright and Moral Rights for the articles on this site are retained by the individual authors and/or other copyright owners. For more information on Open Research Online’s data policy on reuse of materials please consult the policies page. oro.open.ac.uk bs_bs_banner Environmental Microbiology (2015) 17(2), 257–277 doi:10.1111/1462-2920.12598 Minireview Multiplication of microbes below 0.690 water activity: implications for terrestrial and extraterrestrial life Andrew Stevenson,1 Jürgen Burkhardt,2 Summary Charles S. Cockell,3 Jonathan A. Cray,1 Since a key requirement of known life forms is avail- Jan Dijksterhuis,4 Mark Fox-Powell,3 Terence P.