Molecular Mechanisms of Inward and Outward Budding from Multivesicular Endosomes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

View of HER2: Human Epidermal Growth Factor Receptor 2; TNBC: Triple-Negative Breast Resistance to Systemic Therapy in Patients with Breast Cancer
Wen et al. Cancer Cell Int (2018) 18:128 https://doi.org/10.1186/s12935-018-0625-9 Cancer Cell International PRIMARY RESEARCH Open Access Sulbactam‑enhanced cytotoxicity of doxorubicin in breast cancer cells Shao‑hsuan Wen1†, Shey‑chiang Su2†, Bo‑huang Liou3, Cheng‑hao Lin1 and Kuan‑rong Lee1* Abstract Background: Multidrug resistance (MDR) is a major obstacle in breast cancer treatment. The predominant mecha‑ nism underlying MDR is an increase in the activity of adenosine triphosphate (ATP)-dependent drug efux trans‑ porters. Sulbactam, a β-lactamase inhibitor, is generally combined with β-lactam antibiotics for treating bacterial infections. However, sulbactam alone can be used to treat Acinetobacter baumannii infections because it inhibits the expression of ATP-binding cassette (ABC) transporter proteins. This is the frst study to report the efects of sulbactam on mammalian cells. Methods: We used the breast cancer cell lines as a model system to determine whether sulbactam afects cancer cells. The cell viabilities in the present of doxorubicin with or without sulbactam were measured by MTT assay. Protein identities and the changes in protein expression levels in the cells after sulbactam and doxorubicin treatment were determined using LC–MS/MS. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) was used to analyze the change in mRNA expression levels of ABC transporters after treatment of doxorubicin with or without sulbactam. The efux of doxorubicin was measures by the doxorubicin efux assay. Results: MTT assay revealed that sulbactam enhanced the cytotoxicity of doxorubicin in breast cancer cells. The results of proteomics showed that ABC transporter proteins and proteins associated with the process of transcription and initiation of translation were reduced. -

Trafficking Routes to the Plant Vacuole: Connecting Alternative and Classical Pathways
This is a repository copy of Trafficking routes to the plant vacuole: connecting alternative and classical pathways. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/124374/ Version: Accepted Version Article: Di Sansebastiano, GP, Barozzi, F, Piro, G et al. (2 more authors) (2018) Trafficking routes to the plant vacuole: connecting alternative and classical pathways. Journal of Experimental Botany, 69 (1). pp. 79-90. ISSN 0022-0957 https://doi.org/10.1093/jxb/erx376 © The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology. This is an author produced version of a paper published in Journal of Experimental Botany. Uploaded in accordance with the publisher's self-archiving policy. Reuse Items deposited in White Rose Research Online are protected by copyright, with all rights reserved unless indicated otherwise. They may be downloaded and/or printed for private study, or other acts as permitted by national copyright laws. The publisher or other rights holders may allow further reproduction and re-use of the full text version. This is indicated by the licence information on the White Rose Research Online record for the item. Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ 1 Trafficking routes to the Plant Vacuole: 2 connecting alternative and classical pathways. 3 4 Gian Pietro Di Sansebastiano1*, Fabrizio Barozzi1, Gabriella Piro1, Jurgen Denecke2 and 5 Carine de Marcos Lousa2,3 * 6 1.! DiSTeBA (Dipartimento di Scienze e Tecnologie Biologiche ed Ambientali), 7 University of Salento, Campus ECOTEKNE, 73100 Lecce, Italy; 8 [email protected] and [email protected]. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Sorting Nexins in Protein Homeostasis Sara E. Hanley1,And Katrina F
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 6 November 2020 doi:10.20944/preprints202011.0241.v1 Sorting nexins in protein homeostasis Sara E. Hanley1,and Katrina F. Cooper2* 1Department of Molecular Biology, Graduate School of Biomedical Sciences, Rowan University, Stratford, NJ, 08084, USA 1 [email protected] 2 [email protected] * [email protected] Tel: +1 (856)-566-2887 1Department of Molecular Biology, Graduate School of Biomedical Sciences, Rowan University, Stratford, NJ, 08084, USA Abstract: Sorting nexins (SNXs) are a highly conserved membrane-associated protein family that plays a role in regulating protein homeostasis. This family of proteins is unified by their characteristic phox (PX) phosphoinositides binding domain. Along with binding to membranes, this family of SNXs also comprises a diverse array of protein-protein interaction motifs that are required for cellular sorting and protein trafficking. SNXs play a role in maintaining the integrity of the proteome which is essential for regulating multiple fundamental processes such as cell cycle progression, transcription, metabolism, and stress response. To tightly regulate these processes proteins must be expressed and degraded in the correct location and at the correct time. The cell employs several proteolysis mechanisms to ensure that proteins are selectively degraded at the appropriate spatiotemporal conditions. SNXs play a role in ubiquitin-mediated protein homeostasis at multiple levels including cargo localization, recycling, degradation, and function. In this review, we will discuss the role of SNXs in three different protein homeostasis systems: endocytosis lysosomal, the ubiquitin-proteasomal, and the autophagy-lysosomal system. The highly conserved nature of this protein family by beginning with the early research on SNXs and protein trafficking in yeast and lead into their important roles in mammalian systems. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

Direct Lysosome-Based Autophagy of Lipid Droplets in Hepatocytes
Direct lysosome-based autophagy of lipid droplets in hepatocytes Ryan J. Schulzea,b,1, Eugene W. Kruegera,b,1, Shaun G. Wellera,b, Katherine M. Johnsona,b, Carol A. Caseyc, Micah B. Schotta,b, and Mark A. McNivena,b,2 aDepartment of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN 55905; bDivision of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN 55905; and cDepartment of Internal Medicine, University of Nebraska Medical Center, Omaha, NE 68198 Edited by Tobias C. Walther, Harvard School of Public Health, Boston, MA, and accepted by Editorial Board Member Joseph L. Goldstein October 13, 2020 (received for review June 4, 2020) Hepatocytes metabolize energy-rich cytoplasmic lipid droplets (LDs) process that appears to play an especially important role in the in the lysosome-directed process of autophagy. An organelle- degradation of hepatocellular LDs (15). A selective form of LD- selective form of this process (macrolipophagy) results in the engulf- centric autophagy known as lipophagy is thought to involve the ment of LDs within double-membrane delimited structures (auto- recognition of as-of-yet unidentified LD-specific receptors to phagosomes) before lysosomal fusion. Whether this is an promote the localized assembly and extension of a sequestering exclusive autophagic mechanism used by hepatocytes to catabolize phagophore around the perimeter of the LD (16, 17). How this LDs is unclear. It is also unknown whether lysosomes alone might be phagophore is targeted to (and extended around) the LD surface to sufficient to mediate LD turnover in the absence of an autophago- facilitate lipophagy remains unclear. Once fully enclosed, the somal intermediate. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Ubiquitin-Dependent Sorting in Endocytosis
Downloaded from http://cshperspectives.cshlp.org/ on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Ubiquitin-Dependent Sorting in Endocytosis Robert C. Piper1, Ivan Dikic2, and Gergely L. Lukacs3 1Department of Molecular Physiology and Biophysics, University of Iowa, Iowa City, Iowa 52242 2Buchmann Institute for Molecular Life Sciences and Institute of Biochemistry II, Goethe University School of Medicine, D-60590 Frankfurt, Germany 3Department of Physiology, McGill University, Montre´al, Que´bec H3G 1Y6, Canada Correspondence: [email protected] When ubiquitin (Ub) is attached to membrane proteins on the plasma membrane, it directs them through a series of sorting steps that culminate in their delivery to the lumen of the lysosome where they undergo complete proteolysis. Ubiquitin is recognized by a series of complexes that operate at a number of vesicle transport steps. Ubiquitin serves as a sorting signal for internalization at the plasma membrane and is the major signal for incorporation into intraluminal vesicles of multivesicular late endosomes. The sorting machineries that catalyze these steps can bind Ub via a variety of Ub-binding domains. At the same time, many of these complexes are themselves ubiquitinated, thus providing a plethora of potential mechanisms to regulate their activity. Here we provide an overview of how membrane proteins are selected for ubiquitination and deubiquitination within the endocytic pathway and how that ubiquitin signal is interpreted by endocytic sorting machineries. HOW PROTEINS ARE SELECTED FOR distinctions concerning different types of ligas- UBIQUITINATION es, their specificity for forming particular poly- Ub chains, and even the kinds of residues in biquitin (Ub) is covalently attached to sub- substrate proteins that can be ubiquitin modi- Ustrate proteins by the concerted action of fied, have been blurred with the discovery of E2-conjugating enzymes and E3 ligases (Var- mixed poly-Ub chains, new enzymatic mecha- shavsky 2012). -

Retromer-Mediated Endosomal Protein Sorting: the Role of Unstructured Domains ⇑ Aamir S
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Letters 589 (2015) 2620–2626 journal homepage: www.FEBSLetters.org Review Retromer-mediated endosomal protein sorting: The role of unstructured domains ⇑ Aamir S. Mukadam, Matthew N.J. Seaman Cambridge Institute for Medical Research, Dept. of Clinical Biochemistry, University of Cambridge, Wellcome Trust/MRC Building, Addenbrookes Hospital, Cambridge CB2 0XY, United Kingdom article info abstract Article history: The retromer complex is a key element of the endosomal protein sorting machinery that is con- Received 6 May 2015 served through evolution and has been shown to play a role in diseases such as Alzheimer’s disease Revised 21 May 2015 and Parkinson’s disease. Through sorting various membrane proteins (cargo), the function of retro- Accepted 26 May 2015 mer complex has been linked to physiological processes such as lysosome biogenesis, autophagy, Available online 10 June 2015 down regulation of signalling receptors and cell spreading. The cargo-selective trimer of retromer Edited by Wilhelm Just recognises membrane proteins and sorts them into two distinct pathways; endosome-to-Golgi retrieval and endosome-to-cell surface recycling and additionally the cargo-selective trimer func- tions as a hub to recruit accessory proteins to endosomes where they may regulate and/or facilitate Keywords: Retromer retromer-mediated endosomal proteins sorting. Unstructured domains present in cargo proteins or Endosome accessory factors play key roles in both these aspects of retromer function and will be discussed in Sorting this review. FAM21 Ó 2015 Federation of European Biochemical Societies. -
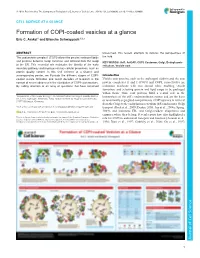
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong.