1/F Noise Modelling and Characterization for CMOS Quanta Image Sensors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CMOS Sensors Enable Phone Cameras, HD Video
CMOS Sensors Enable Phone Cameras, HD Video NASA Technology to come of age by the late 1980s. These image sensors already used in CCDs. Using this technique, he measured comprise an array of photodetecting pixels that collect a pixel’s voltage both before and after an exposure. “It’s eople told me, ‘You’re an idiot to work on charges when exposed to light and transfer those charges, like when you go to the deli counter, and they weigh the this,’” Eric Fossum recalls of his early experi- pixel to pixel, to the corner of the array, where they are container, then weigh it again with the food,” he explains. ments with what was at the time an alternate “P amplified and measured. The sampling corrected for the slight thermal charges and form of digital image sensor at NASA’s Jet Propulsion While CCD sensors are capable of producing scientific- transistor fluctuations that are latent in photodetector Laboratory (JPL). grade images, though, they require a lot of power and readout, and it resulted in a clearer image. His invention of the complementary metal oxide extremely high charge-transfer efficiency. These difficulties Because CMOS pixels are signal amplifiers themselves, semiconductor (CMOS) image sensor would go on to are compounded when the number of pixels is increased for they can each read out their own signals, rather than trans- become the Space Agency’s single most ubiquitous spinoff higher resolution or when video frame rates are sped up. ferring all the charges to a single amplifier. This lowered technology, dominating the digital imaging industries and Fossum was an expert in CCD technology—it was why voltage requirements and eliminated charge transfer- enabling cell phone cameras, high-definition video, and JPL hired him in 1990—but he believed he could make efficiency issues. -

Modeling for Ultra Low Noise CMOS Image Sensors
Thèse n° 7661 Modeling for Ultra Low Noise CMOS Image Sensors Présentée le 10 septembre 2020 à la Faculté des sciences et techniques de l’ingénieur Laboratoire de circuits intégrés Programme doctoral en microsystèmes et microélectronique pour l’obtention du grade de Docteur ès Sciences par Raffaele CAPOCCIA Acceptée sur proposition du jury Prof. E. Charbon, président du jury Prof. C. Enz, directeur de thèse Prof. A. Theuwissen, rapporteur Prof. E. Fossum, rapporteur Dr J.-M. Sallese, rapporteur 2020 Ma Nino non aver paura di sbagliare un calcio di rigore, non è mica da questi particolari che si giudica un giocatore, un giocatore lo vedi dal coraggio, dall’altruismo e dalla fantasia. — Francesco De Gregori To my family. Acknowledgements I would like to express my deep gratitude to my thesis advisor, Prof. Christian Enz. His guid- ance and support over the past few years helped me to expand my knowledge in the area of device modeling and sensor design. I am genuinely grateful for having taught me how important is to be methodical and to insist on the detail. My grateful thanks are also extended to Dr. Assim Boukhayma for his contribution and for having introduced me to the fascinating world of image sensors. His help and encouragement over the entire time of my Ph.D. studies daily motivated me to improve the quality of the research. I wish to thank Dr. Farzan Jazaeri for everything he taught me during my Ph.D. His passion for physics and device modeling has been a source of inspiration for the research. I would also like to express my deepest gratitude to Prof. -

CMOS Active Pixel Image Sensors for Highly Integrated Imaging Systems
IEEE JOURNAL OF SOLID-STATE CIRCUITS, VOL. 32, NO. 2, FEBRUARY 1997 187 CMOS Active Pixel Image Sensors for Highly Integrated Imaging Systems Sunetra K. Mendis, Member, IEEE, Sabrina E. Kemeny, Member, IEEE, Russell C. Gee, Member, IEEE, Bedabrata Pain, Member, IEEE, Craig O. Staller, Quiesup Kim, Member, IEEE, and Eric R. Fossum, Senior Member, IEEE Abstract—A family of CMOS-based active pixel image sensors Charge-coupled devices (CCD’s) are currently the dominant (APS’s) that are inherently compatible with the integration of on- technology for image sensors. CCD arrays with high fill-factor, chip signal processing circuitry is reported. The image sensors small pixel sizes, and large formats have been achieved and were fabricated using commercially available 2-"m CMOS pro- cesses and both p-well and n-well implementations were explored. some signal processing operations have been demonstrated The arrays feature random access, 5-V operation and transistor- with charge-domain circuits [1]–[3]. However, CCD’s cannot transistor logic (TTL) compatible control signals. Methods of be easily integrated with CMOS circuits due to additional on-chip suppression of fixed pattern noise to less than 0.1% fabrication complexity and increased cost. Also, CCD’s are saturation are demonstrated. The baseline design achieved a pixel high capacitance devices so that on-chip CMOS drive electron- size of 40 "m 40 "m with 26% fill-factor. Array sizes of 28 28 elements and 128 128 elements have been fabricated and ics would dissipate prohibitively high power levels for large characterized. Typical output conversion gain is 3.7 "V/e for the area arrays (2–3 W). -

Fundamental Sensitivity Limit Imposed by Dark 1/F Noise in the Low Optical Signal Detection Regime
Fundamental sensitivity limit imposed by dark 1/f noise in the low optical signal detection regime Emily J. McDowell1, Jian Ren2, and Changhuei Yang1,2 1Department of Bioengineering, 2Department of Electrical Engineering, Mail Code 136-93,California Institute of Technology, Pasadena, CA, 91125 [email protected] Abstract: The impact of dark 1/f noise on fundamental signal sensitivity in direct low optical signal detection is an understudied issue. In this theoretical manuscript, we study the limitations of an idealized detector with a combination of white noise and 1/f noise, operating in detector dark noise limited mode. In contrast to white noise limited detection schemes, for which there is no fundamental minimum signal sensitivity limit, we find that the 1/f noise characteristics, including the noise exponent factor and the relative amplitudes of white and 1/f noise, set a fundamental limit on the minimum signal that such a detector can detect. © 2008 Optical Society of America OCIS codes: (230.5160) Photodetectors; (040.3780) Low light level References and links 1. S. O. Flyckt and C. Marmonier, Photomultiplier tubes: Principles and applications (Photonis, 2002). 2. M. C. Teich, K. Matsuo, and B. E. A. Saleh, "Excess noise factors for conventional and superlattice avalanche photodiodes and photomultiplier tubes," IEEE J. Quantum Electron. 22, 1184-1193 (1986). 3. E. J. McDowell, X. Cui, Y. Yaqoob, and C. Yang, "A generalized noise variance analysis model and its application to the characterization of 1/f noise," Opt. Express 15, (2007). 4. P. Dutta and P. M. Horn, "Low-frequency fluctuations in solids - 1/f noise," Rev. -

Print Special Issue Flyer
IMPACT FACTOR 3.576 an Open Access Journal by MDPI Photon-Counting Image Sensors Guest Editors: Message from the Guest Editors Prof. Dr. Eric Fossum Dear Colleagues, [email protected] The field of photon-counting image sensors is advancing Prof. Dr. Nobukazu Teranishi rapidly with the development of various solid-state image [email protected] sensor technologies including single photon avalanche Prof. Dr. Albert Theuwissen detectors (SPADs) and deep-sub-electron read noise CMOS [email protected] image sensor pixels. This foundational platform technology will enable opportunities for new imaging modalities and Dr. David Stoppa instrumentation for science and industry, as well as new [email protected] consumer applications. Papers discussing various photon- Prof. Dr. Edoardo Charbon counting image sensor technologies and selected new [email protected] applications are presented in this all-invited Special Issue. Prof. Dr. Eric R. Fossum Prof. Dr. Edoardo Charbon Deadline for manuscript Dr. David Stoppa submissions: closed (12 February 2016) Prof. Dr. Nobukazu Teranishi Prof. Dr. Albert Theuwissen Guest Editors mdpi.com/si/6076 SpeciaIslsue IMPACT FACTOR 3.576 an Open Access Journal by MDPI Editor-in-Chief Message from the Editor-in-Chief Prof. Dr. Vittorio M.N. Passaro Sensors is a leading journal devoted to fast publication of Dipartimento di Ingegneria the latest achievements of technological developments Elettrica e dell'Informazione and scientific research in the huge area of physical, (Department of Electrical and Information Engineering), chemical and biochemical sensors, including remote Politecnico di Bari, Via Edoardo sensing and sensor networks. Both experimental and Orabona n. -

Boundary-Preserved Deep Denoising of the Stochastic Resonance Enhanced Multiphoton Images
Boundary-Preserved Deep Denoising of the Stochastic Resonance Enhanced Multiphoton Images SHENG-YONG NIU1,2, LUN-ZHANG GUO3, YUE LI4, TZUNG-DAU WANG5, YU TSAO1,*, TZU-MING LIU4,* 1. Research Center for Information Technology Innovation (CITI), Academia Sinica, Taipei, Taiwan. 2. Department of Computer Science and Engineering, University of California San Diego, CA, USA. 3. Institute of Biomedical Engineering, National Taiwan University, Taipei 10617, Taiwan. 4. Institute of Translational Medicine, Faculty of Health Sciences, University of Macau, Macao SAR, China. 5. Cardiovascular Center and Division of Cardiology, Department of Internal Medicine, National Taiwan University Hospital and College of Medicine, Taipei 10002, Taiwan. * [email protected] and [email protected] Abstract: As the rapid growth of high-speed and deep-tissue imaging in biomedical research, it is urgent to find a robust and effective denoising method to retain morphological features for further texture analysis and segmentation. Conventional denoising filters and models can easily suppress perturbative noises in high contrast images. However, for low photon budget multi- photon images, high detector gain will not only boost signals, but also bring huge background noises. In such stochastic resonance regime of imaging, sub-threshold signals may be detectable with the help of noises. Therefore, a denoising filter that can smartly remove noises without sacrificing the important cellular features such as cell boundaries is highly desired. In this paper, we propose a convolutional neural network based denoising autoencoder method, Fully Convolutional Deep Denoising Autoencoder (DDAE), to improve the quality of Three-Photon Fluorescence (3PF) and Third Harmonic Generation (THG) microscopy images. The average of the acquired 200 images of a given location served as the low-noise answer for DDAE training. -
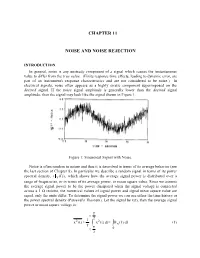
Chapter 11 Noise and Noise Rejection
CHAPTER 11 NOISE AND NOISE REJECTION INTRODUCTION In general, noise is any unsteady component of a signal which causes the instantaneous value to differ from the true value. (Finite response time effects, leading to dynamic error, are part of an instrument's response characteristics and are not considered to be noise.) In electrical signals, noise often appears as a highly erratic component superimposed on the desired signal. If the noise signal amplitude is generally lower than the desired signal amplitude, then the signal may look like the signal shown in Figure 1. Figure 1: Sinusoidal Signal with Noise. Noise is often random in nature and thus it is described in terms of its average behavior (see the last section of Chapter 8). In particular we describe a random signal in terms of its power spectral density, (x (f )) , which shows how the average signal power is distributed over a range of frequencies, or in terms of its average power, or mean square value. Since we assume the average signal power to be the power dissipated when the signal voltage is connected across a 1 Ω resistor, the numerical values of signal power and signal mean square value are equal, only the units differ. To determine the signal power we can use either the time history or the power spectral density (Parseval's Theorem). Let the signal be x(t), then the average signal power or mean square voltage is: T t 2 221 x(t) x(t)dtx (f)df (1) T T t 0 2 11-2 Note: the bar notation, , denotes a time average taken over many oscillations of the signal. -

Grant County Planning and Zoning on WES
Bald and Golden Eagles, Grant and Codington Counties, 2017 & 2018 Grant County Planning and Zoning on WES April 17th, 2018 1. Questions from the public 2. General information and where to find more information, includes setback information from Vesta manuals, health, decommissioning and return on Tax Production Credit 3. Health information and where to find more, includes Cooper interview and personal testimony 4. Property value loses, includes report from PUC debunking the Next Era claim 5. Decommissioning issues 6. Language used in Wind Contracts This book contains websites and where to find more information. If you have questions, need additional reports you may contact Vince Meyer 605-949-1916 Facts, Findings and Conclusions We would like FINDINGS to the following issues to be addressed as answers in local newspapers for the public to review and discuss. 1. Has Next Era, APEX, or any wind energy developer, provided scientific methods to prove flicker, sound, infrasound or other nuisance, will trespass on to non-participants' land, for a certain allotment of time, please present the methods and studies to support the claim. 2. Have Planning and Zoning members and Commissioners read a Turbine manual for all the models being proposed to be used for safety setbacks? If so, where is a manual for the public to evaluate? 3. Has a MSDS for the county? 4. Wind farms are privately owned. They do not supply electricity to the public. Their electricity is for sale. Hence they are not a public utility. How will the county or township handle trespassing on private land be it rights-of-ways or other types of property? 5. -
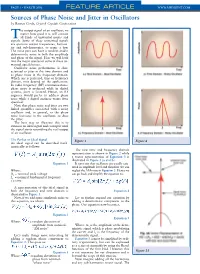
Sources of Phase Noise and Jitter in Oscillators by Ramon Cerda, Crystek Crystals Corporation
PAGE • MARCH 2006 FEATURE ARTICLE WWW.MPDIGEST.COM Sources of Phase Noise and Jitter in Oscillators by Ramon Cerda, Crystek Crystals Corporation he output signal of an oscillator, no matter how good it is, will contain Tall kinds of unwanted noises and signals. Some of these unwanted signals are spurious output frequencies, harmon- ics and sub-harmonics, to name a few. The noise part can have a random and/or deterministic noise in both the amplitude and phase of the signal. Here we will look into the major sources of some of these un- wanted signals/noises. Oscillator noise performance is char- acterized as jitter in the time domain and as phase noise in the frequency domain. Which one is preferred, time or frequency domain, may depend on the application. In radio frequency (RF) communications, phase noise is preferred while in digital systems, jitter is favored. Hence, an RF engineer would prefer to address phase noise while a digital engineer wants jitter specified. Note that phase noise and jitter are two linked quantities associated with a noisy oscillator and, in general, as the phase noise increases in the oscillator, so does the jitter. The best way to illustrate this is to examine an ideal signal and corrupt it until the signal starts resembling the real output of an oscillator. The Perfect or Ideal Signal Figure 1 Figure 2 An ideal signal can be described math- ematically as follows: The new time and frequency domain representation is shown in Figure 2 while a vector representation of Equation 3 is illustrated in Figure 3 (a and b.) Equation 1 It turns out that oscillators are usually satu- rated in amplitude level and therefore we can Where: neglect the AM noise in Equation 3. -

NASA Technologies Benefit Society
National Aeronautics and Space Administration NASA TECH N OLOGIE S BE N EFI T SOCIE T Y 2010 spinoff SPINOFF Office of the Chief Technologist On the cover: During the STS-128 space shuttle mission, a space-walking astronaut took this photograph of a portion of the International Space Station (ISS) flying high above Earth’s glowing horizon. This year marks the 10th 2010 anniversary of the ISS, and is also a year of beneficial spinoff technologies, as highlighted by the smaller images lined up against the backdrop of space. Spinoff Program Office NASA Center for AeroSpace Information Daniel Lockney, Editor Bo Schwerin, Senior Writer Lisa Rademakers, Writer John Jones, Graphic Designer Deborah Drumheller, Publications Specialist Table of Contents 7 Foreword 9 Introduction 10 International Space Station Spinoffs 20 Executive Summary 30 NASA Technologies Enhance Our Lives on Earth 32 NASA Partnerships Across the Nation 34 NASA Technologies Benefiting Society Health and Medicine Burnishing Techniques Strengthen Hip Implants ...............................................................38 Signal Processing Methods Monitor Cranial Pressure ..........................................................40 Ultraviolet-Blocking Lenses Protect, Enhance Vision ..........................................................42 Hyperspectral Systems Increase Imaging Capabilities ..........................................................44 Transportation Programs Model the Future of Air Traffic Management ......................................................48 -

MT-048: Op Amp Noise Relationships
MT-048 TUTORIAL Op Amp Noise Relationships: 1/f Noise, RMS Noise, and Equivalent Noise Bandwidth "1/f" NOISE The general characteristic of op amp current or voltage noise is shown in Figure 1 below. NOISE 3dB/Octave 1 nV / √Hz e , i = k F n n C f or μV / √Hz 1 CORNER f en, in WHITE NOISE k F LOG f C Figure 1: Frequency Characteristic of Op Amp Noise At high frequencies the noise is white (i.e., its spectral density does not vary with frequency). This is true over most of an op amp's frequency range, but at low frequencies the noise spectral density rises at 3 dB/octave, as shown in Figure 1 above. The power spectral density in this region is inversely proportional to frequency, and therefore the voltage noise spectral density is inversely proportional to the square root of the frequency. For this reason, this noise is commonly referred to as 1/f noise. Note however, that some textbooks still use the older term flicker noise. The frequency at which this noise starts to rise is known as the 1/f corner frequency (FC) and is a figure of merit—the lower it is, the better. The 1/f corner frequencies are not necessarily the same for the voltage noise and the current noise of a particular amplifier, and a current feedback op amp may have three 1/f corners: for its voltage noise, its inverting input current noise, and its non-inverting input current noise. The general equation which describes the voltage or current noise spectral density in the 1/f region is 1 = Fk,i,e , Eq. -

Flicker Noise of Phase in Rf Amplifiers and Frequency Multipliers
FLICKER NOISE OF PHASE IN RF AMPLIFIERS AND FREQUENCYMULTIPLIERS: CHARACTERIZATION, CAUSE, AND CURE Donald Halford, A. E.Wainwright, and James A. Barnes Time and Frequency Division National Bureau of Standards, Boulder, Colorado 80302 Summary The high phase stability of atomic frequency standards has called for the development of associated electronic equipment of equivalent or superior stability. We have surveyed the performance of existing high quality frequency multipliers and RF amplifiers which operate in the range of 5 MHz to microwave frequencies. We were most interested in the phase noise in the range of Fourierfrequencies, f, of about Hz to lot3 Hz, sincemost electronic servo systems in existing atomic frequency standards use modula- tion frequencies which fall within this range. This range also includes the passive linewidths of existing atomic frequency standards. We found that all state-of-the-art, solid state amplifiers and frequency multipliers in our survey had random phase fluctuations (phase noise) of significant intensity and witha spectral density proportional to l/f (flicker noise of phase). Surprisingly, the flicker noise of phase was found to be approximately the same in most of the apparatus included in the survey. The typical one-sided spectral density of the phase noise was about (10- radian2)/f when referred to the input frequency, and the best performance was only 6 dB better. This performance was independent of the input fre- quency over at least the surveyed range of 5 MHz to 100 MHz, and did not depend upon the multiplication factor. This noise level exceeds by about 20 dB the stability which is needed for full compatibility with existing hydro- gen atom masers and with proposed high flux cesium beam designs.