2016 Phi Zeta Proceedings (PDF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

2012 Phi Zeta Proceedings
The Honor Society of Veterinary Medicine Epsilon Chapter November 7, 2012 Research Emphasis Day AUBURN UNIVERSITY COLLEGE OF VETERINARY MEDICINE PHI ZETA EPSILON CHAPTER COLLEGE OF VETERINARY MEDICINE AUBURN UNIVERSITY welcomes you to our PHI ZETA RESEARCH DAY FORUM November 7, 2012 We want to thank all the presenters, their co-investigators and mentors for their participation in this annual event. We also want to thank all sponsors for their generous support without which this event would not be possible: Department of Anatomy, Physiology, and Pharmacology Department of Clinical Sciences Department of Pathobiology Office of the Assoc. Dean for Research and Graduate Studies Office of the Associate Dean of Academic Affairs Office of the Dean PROGRAM PHI ZETA RESEARCH EMPHASIS DAY November 7, 2012 – Overton-Goodwin Center Graduate Student Platform Presentations (OEW) OEW 248 OEW 251 8:30 Amy Back 8:45 Allison Bradbury 8:45 Michelle Aono 9:00 Erfan Chowdhury 9:00 Heather Davis 9:15 Elaine Norton 9:15 Elizabeth Barrett 9:30 Fernanda Cesar 9:30 Hui Huang 9:45 Break 9:45 Break 10:00 Anil Poudel 10:00 Valeria Albanese 10:15 India Napier 10:15 Kh. S. Rahman 10:30 Rucha Gurjar 10:30 Margaret Salter 10:45 Victoria McCurdy 10:45 Wes Campbell 11:00-1:00 Poster Presentations- Overton Education Wing (Poster Session Presenters are present 11:00 – 12:00) Veterinary Student Platform Presentation (OEW 248) 1:30 Jeremy Fleming 1:45 Amelia Nuwer 2:00 Hannah Findlay 2:15 Kelsie Theis 2:30 Nikki McAdams 1 PROGRAM Post-graduate/Faculty Platform Presentations (OEW 248) 2:45 Deepa Bedi 3:00 Heather Gray-Edwards 3:15 Merrilee Holland 3:30 Jeremy Foote 3:45 Snack Break – Joy Goodwin Cafeteria 4:00 Keynote Lecture – Overton Auditorium – Dr. -

The Mysterious Orphans of Mycoplasmataceae
The mysterious orphans of Mycoplasmataceae Tatiana V. Tatarinova1,2*, Inna Lysnyansky3, Yuri V. Nikolsky4,5,6, and Alexander Bolshoy7* 1 Children’s Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, 90027, California, USA 2 Spatial Science Institute, University of Southern California, Los Angeles, 90089, California, USA 3 Mycoplasma Unit, Division of Avian and Aquatic Diseases, Kimron Veterinary Institute, POB 12, Beit Dagan, 50250, Israel 4 School of Systems Biology, George Mason University, 10900 University Blvd, MSN 5B3, Manassas, VA 20110, USA 5 Biomedical Cluster, Skolkovo Foundation, 4 Lugovaya str., Skolkovo Innovation Centre, Mozhajskij region, Moscow, 143026, Russian Federation 6 Vavilov Institute of General Genetics, Moscow, Russian Federation 7 Department of Evolutionary and Environmental Biology and Institute of Evolution, University of Haifa, Israel 1,2 [email protected] 3 [email protected] 4-6 [email protected] 7 [email protected] 1 Abstract Background: The length of a protein sequence is largely determined by its function, i.e. each functional group is associated with an optimal size. However, comparative genomics revealed that proteins’ length may be affected by additional factors. In 2002 it was shown that in bacterium Escherichia coli and the archaeon Archaeoglobus fulgidus, protein sequences with no homologs are, on average, shorter than those with homologs [1]. Most experts now agree that the length distributions are distinctly different between protein sequences with and without homologs in bacterial and archaeal genomes. In this study, we examine this postulate by a comprehensive analysis of all annotated prokaryotic genomes and focusing on certain exceptions. -

Ramiro Isaza, DVM, MS, MPH, DACZM (Updated October 29, 2013)
Curriculum Vitae Ramiro Isaza, DVM, MS, MPH, DACZM (Updated October 29, 2013) PRESENT POSITION: Associate Professor of Zoological Medicine (Graduate Faculty Member) CONTACT INFORMATION: ACADEMIC ADDRESS Department of Small Animal Clinical Sciences College of Veterinary Medicine PO Box 100126 University of Florida Gainesville, Florida 32610-0126 TELEPHONE (352) 294-4443 FAX (352) 392-6125 E-MAIL [email protected] EDUCATION: MPH 2011 College of Public Health and Health Professions University of Florida Gainesville, Florida MS 1993 College of Veterinary Medicine University of Florida Gainesville, Florida DVM 1988 New York State College of Veterinary Medicine Cornell University Ithaca, New York BS 1983 Zoology, with High Honors University of Florida Gainesville, Florida AA/AS 1979 Zoo Keeper Training Program Santa Fe Community College Gainesville, Florida POST-GRADUATE TRAINING IN ZOOLOGICAL MEDICINE: Residency 1992 Wildlife and Zoological Medicine College of Veterinary Medicine University of Florida Gainesville, Florida Internship 1990 Zoological Medicine Riverbanks Zoological Park Columbia, SC Ramiro Isaza Page 2 BOARD CERTIFICATION: Diplomate (#40 / 127) 1996 American College of Zoological Medicine VETERINARY LICENSURES: Florida, California APPOINTMENTS AND PROFESSIONAL EXPERIENCE: 2007-present Associate Professor Zoological Medicine Service Department of Small Animal Clinical Sciences College of Veterinary Medicine, University of Florida Gainesville, Florida 2003-2007 Assistant Professor Zoological Medicine Service Department of Small Animal -

Role of Protein Phosphorylation in Mycoplasma Pneumoniae
Pathogenicity of a minimal organism: Role of protein phosphorylation in Mycoplasma pneumoniae Dissertation zur Erlangung des mathematisch-naturwissenschaftlichen Doktorgrades „Doctor rerum naturalium“ der Georg-August-Universität Göttingen vorgelegt von Sebastian Schmidl aus Bad Hersfeld Göttingen 2010 Mitglieder des Betreuungsausschusses: Referent: Prof. Dr. Jörg Stülke Koreferent: PD Dr. Michael Hoppert Tag der mündlichen Prüfung: 02.11.2010 “Everything should be made as simple as possible, but not simpler.” (Albert Einstein) Danksagung Zunächst möchte ich mich bei Prof. Dr. Jörg Stülke für die Ermöglichung dieser Doktorarbeit bedanken. Nicht zuletzt durch seine freundliche und engagierte Betreuung hat mir die Zeit viel Freude bereitet. Des Weiteren hat er mir alle Freiheiten zur Verwirklichung meiner eigenen Ideen gelassen, was ich sehr zu schätzen weiß. Für die Übernahme des Korreferates danke ich PD Dr. Michael Hoppert sowie Prof. Dr. Heinz Neumann, PD Dr. Boris Görke, PD Dr. Rolf Daniel und Prof. Dr. Botho Bowien für das Mitwirken im Thesis-Komitee. Der Studienstiftung des deutschen Volkes gilt ein besonderer Dank für die finanzielle Unterstützung dieser Arbeit, durch die es mir unter anderem auch möglich war, an Tagungen in fernen Ländern teilzunehmen. Prof. Dr. Michael Hecker und der Gruppe von Dr. Dörte Becher (Universität Greifswald) danke ich für die freundliche Zusammenarbeit bei der Durchführung von zahlreichen Proteomics-Experimenten. Ein ganz besonderer Dank geht dabei an Katrin Gronau, die mich in die Feinheiten der 2D-Gelelektrophorese eingeführt hat. Außerdem möchte ich mich bei Andreas Otto für die zahlreichen Proteinidentifikationen in den letzten Monaten bedanken. Nicht zu vergessen ist auch meine zweite Außenstelle an der Universität in Barcelona. Dr. Maria Lluch-Senar und Dr. -

Genomic Islands in Mycoplasmas
G C A T T A C G G C A T genes Review Genomic Islands in Mycoplasmas Christine Citti * , Eric Baranowski * , Emilie Dordet-Frisoni, Marion Faucher and Laurent-Xavier Nouvel Interactions Hôtes-Agents Pathogènes (IHAP), Université de Toulouse, INRAE, ENVT, 31300 Toulouse, France; [email protected] (E.D.-F.); [email protected] (M.F.); [email protected] (L.-X.N.) * Correspondence: [email protected] (C.C.); [email protected] (E.B.) Received: 30 June 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Bacteria of the Mycoplasma genus are characterized by the lack of a cell-wall, the use of UGA as tryptophan codon instead of a universal stop, and their simplified metabolic pathways. Most of these features are due to the small-size and limited-content of their genomes (580–1840 Kbp; 482–2050 CDS). Yet, the Mycoplasma genus encompasses over 200 species living in close contact with a wide range of animal hosts and man. These include pathogens, pathobionts, or commensals that have retained the full capacity to synthesize DNA, RNA, and all proteins required to sustain a parasitic life-style, with most being able to grow under laboratory conditions without host cells. Over the last 10 years, comparative genome analyses of multiple species and strains unveiled some of the dynamics of mycoplasma genomes. This review summarizes our current knowledge of genomic islands (GIs) found in mycoplasmas, with a focus on pathogenicity islands, integrative and conjugative elements (ICEs), and prophages. Here, we discuss how GIs contribute to the dynamics of mycoplasma genomes and how they participate in the evolution of these minimal organisms. -

Student Handbook for Students in the Professional Veterinary Program
2021-22 University of Florida College of Veterinary Medicine Student Handbook For Students in the Professional Veterinary Program Student Handbook 2021-2022 UF reserves the right to implement new regulations and policies not currently included in this document. The university will make a reasonable attempt to inform students of changes in regulations or policies. The most updated version of the student handbook can be found at: http://education.vetmed.ufl.edu/dvm-curriculum/student-handbook/ Contents I. Introduction ........................................................................................................................... 6 Our Mission ......................................................................................................................................................................... 6 Calendar ............................................................................................................................................................................... 6 II Resources ................................................................................................................................ 7 Alumni Affairs ..................................................................................................................................................................... 7 Auditing a Course ................................................................................................................................................................ 7 Accessing Cornerstone from Home .................................................................................................................................... -

Genetic Profiling of Mycoplasma Hyopneumoniae Melissa L
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2005 Genetic profiling of Mycoplasma hyopneumoniae Melissa L. Madsen Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Microbiology Commons, Molecular Biology Commons, and the Veterinary Medicine Commons Recommended Citation Madsen, Melissa L., "Genetic profiling of Mycoplasma hyopneumoniae " (2005). Retrospective Theses and Dissertations. 1793. https://lib.dr.iastate.edu/rtd/1793 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. NOTE TO USERS This reproduction is the best copy available. ® UMI Genetic profiling of Mycoplasma hyopneumoniae by Melissa L. Madsen A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Molecular, Cellular and Developmental Biology Program of Study Committee: F. Chris Minion, Major Professor Daniel S. Nettleton Gregory J. Phillips Eileen L. Thacker Eve Wurtele Iowa State University Ames, Iowa 2005 UMI Number: 3200480 INFORMATION TO USERS The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleed-through, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. -

Ian Keith Hawkins Present Rank: Assistant
Curriculum Vitae Ian K. Hawkins 1. ACADEMIC HISTORY AND PROFESSIONAL EXPERIENCE: Name: Ian Keith Hawkins Present Rank: Assistant Professor Proportion of Time Assignment: 80% Service 20% Research Tenure Status: Not tenured Administrative Title: Assistant Professor and Veterinary Pathologist (TVDIL 2015-present). Graduate Faculty Status: Not a member Highest Degrees: Doctor of Veterinary Medicine University of Missouri May 2007 Bachelor of Science, Zoology and Entomology The Ohio State University June 2003 Specialty Training and Certification: Diplomate American College of Veterinary Pathologists September 2011 Anatomic Pathology Residency University of Florida July 2010 Professional Licensure: Georgia Veterinary Medical License, Faculty License, 2015 – present USDA Accreditation, Category II Veterinarian, 2015 – present Diplomate, American College of Veterinary Pathologists, 2011 – present Royal College of Veterinary Surgeons, 2011 – present Ohio Veterinary Medical License, 2007 – present Academic Positions: 2015-present: Assistant Professor and Veterinary Pathologist, University of Georgia, College of Veterinary Medicine, Tifton Veterinary Diagnostic and Investigational Laboratory. Professional Positions: 2011-2015: Veterinary Pathologist, Bridge Pathology Limited, Bristol, United Kingdom. 2011: Veterinarian, Lindquist Veterinary Center, Kirksville, Missouri. 2007-2010: Anatomic Pathology Resident, University of Florida, College of Veterinary Medicine, Department of Infectious Diseases and Pathology, Gainesville, Florida. Awards and Scholarships: -

Comparative Analyses of Whole-Genome Protein Sequences
www.nature.com/scientificreports OPEN Comparative analyses of whole- genome protein sequences from multiple organisms Received: 7 June 2017 Makio Yokono 1,2, Soichirou Satoh3 & Ayumi Tanaka1 Accepted: 16 April 2018 Phylogenies based on entire genomes are a powerful tool for reconstructing the Tree of Life. Several Published: xx xx xxxx methods have been proposed, most of which employ an alignment-free strategy. Average sequence similarity methods are diferent than most other whole-genome methods, because they are based on local alignments. However, previous average similarity methods fail to reconstruct a correct phylogeny when compared against other whole-genome trees. In this study, we developed a novel average sequence similarity method. Our method correctly reconstructs the phylogenetic tree of in silico evolved E. coli proteomes. We applied the method to reconstruct a whole-proteome phylogeny of 1,087 species from all three domains of life, Bacteria, Archaea, and Eucarya. Our tree was automatically reconstructed without any human decisions, such as the selection of organisms. The tree exhibits a concentric circle-like structure, indicating that all the organisms have similar total branch lengths from their common ancestor. Branching patterns of the members of each phylum of Bacteria and Archaea are largely consistent with previous reports. The topologies are largely consistent with those reconstructed by other methods. These results strongly suggest that this approach has sufcient taxonomic resolution and reliability to infer phylogeny, from phylum to strain, of a wide range of organisms. Te reconstruction of phylogenetic trees is a powerful tool for understanding organismal evolutionary processes. Molecular phylogenetic analysis using ribosomal RNA (rRNA) clarifed the phylogenetic relationship of the three domains, bacterial, archaeal, and eukaryotic1. -

1 Supplementary Material a Major Clade of Prokaryotes with Ancient
Supplementary Material A major clade of prokaryotes with ancient adaptations to life on land Fabia U. Battistuzzi and S. Blair Hedges Data assembly and phylogenetic analyses Protein data set: Amino acid sequences of 25 protein-coding genes (“proteins”) were concatenated in an alignment of 18,586 amino acid sites and 283 species. These proteins included: 15 ribosomal proteins (RPL1, 2, 3, 5, 6, 11, 13, 16; RPS2, 3, 4, 5, 7, 9, 11), four genes (RNA polymerase alpha, beta, and gamma subunits, Transcription antitermination factor NusG) from the functional category of Transcription, three proteins (Elongation factor G, Elongation factor Tu, Translation initiation factor IF2) of the Translation, Ribosomal Structure and Biogenesis functional category, one protein (DNA polymerase III, beta subunit) of the DNA Replication, Recombination and repair category, one protein (Preprotein translocase SecY) of the Cell Motility and Secretion category, and one protein (O-sialoglycoprotein endopeptidase) of the Posttranslational Modification, Protein Turnover, Chaperones category, as annotated in the Cluster of Orthologous Groups (COG) (Tatusov et al. 2001). After removal of multiple strains of the same species, GBlocks 0.91b (Castresana 2000) was applied to each protein in the concatenation to delete poorly aligned sites (i.e., sites with gaps in more than 50% of the species and conserved in less than 50% of the species) with the following parameters: minimum number of sequences for a conserved position: 110, minimum number of sequences for a flank position: 110, maximum number of contiguous non-conserved positions: 32000, allowed gap positions: with half. The signal-to-noise ratio was determined by altering the “minimum length of a block” parameter. -
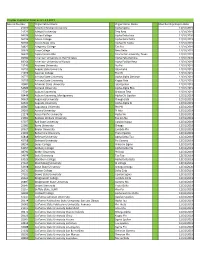
12.3.2019 LPH Chapter Expiration Dates.Xlsx
Chapter Expiration Dates as of 12.3.2019 Record Number Organization Name Organization Name Memberships Expire Date 21070 Abilene Christian University Alpha Sigma 12/31/2019 21523 Adelphi University Zeta Beta 12/31/2019 64336 Adrian College Alpha Delta Iota 12/31/2020 56196 Albion College Alpha Beta Delta 12/31/2019 66999 Alcorn State Univ Alpha Chi Alpha 12/31/2019 34827 Allegheny College Tau Eta 12/31/2019 20626 Alma College Beta Delta 12/31/2019 96670 Alpha Epsilon Rho Concordia University, Texas 12/31/2019 99780 American University in the Emirates Alpha Zeta Gamma 12/31/2020 69264 American University of Kuwait Alpha Epsilon Beta 12/31/2019 23243 Andrews University Nu Psi 12/31/2019 26793 Angelo State University Iota Alpha 12/31/2019 21079 Aquinas College Eta Chi 12/31/2019 26777 Arizona State University Alpha Alpha Omicron 12/31/2019 26799 Arizona State University Kappa Zeta 12/31/2019 20681 Arkansas State University Iota Upsilon 12/31/2019 54689 Ashland University Alpha Alpha Rho 12/31/2019 27741 Auburn University Omicron Zeta 12/31/2019 68446 Auburn University, Montgomery Alpha Chi Upsilon 12/31/2019 50629 Augsburg University Omega Zeta 12/31/2019 42631 Augusta University Alpha Alpha Xi 12/31/2019 40087 Augustana University Phi Phi 12/31/2019 29103 Aurora University Pi Iota 12/31/2019 21178 Azusa Pacific University Alpha Nu 12/31/2019 21061 Baldwin Wallace University Epsilon Nu 12/31/2019 37077 Ball State University Upsilon Kappa 12/31/2019 22981 Barry University Omega 12/31/2019 20627 Baylor University Lambda Phi 12/31/2019 21202 -

Effectors of Mycoplasmal Virulence 1 2 Virulence Effectors of Pathogenic Mycoplasmas 3 4 Meghan A. May1 And
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 27 September 2018 doi:10.20944/preprints201809.0533.v1 1 Running title: Effectors of mycoplasmal virulence 2 3 Virulence Effectors of Pathogenic Mycoplasmas 4 5 Meghan A. May1 and Daniel R. Brown2 6 7 1Department of Biomedical Sciences, College of Osteopathic Medicine, University of New 8 England, Biddeford ME, USA; 2Department of Infectious Diseases and Immunology, College 9 of Veterinary Medicine, University of Florida, Gainesville FL, USA 10 11 Corresponding author: 12 Daniel R. Brown 13 Department of Infectious Diseases and Immunology, College of Veterinary Medicine, 14 University of Florida, Gainesville FL, USA 15 Tel: +1 352 294 4004 16 Email: [email protected] 1 © 2018 by the author(s). Distributed under a Creative Commons CC BY license. Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 27 September 2018 doi:10.20944/preprints201809.0533.v1 17 Abstract 18 Members of the genus Mycoplasma and related organisms impose a substantial burden of 19 infectious diseases on humans and animals, but the last comprehensive review of 20 mycoplasmal pathogenicity was published 20 years ago. Post-genomic analyses have now 21 begun to support the discovery and detailed molecular biological characterization of a 22 number of specific mycoplasmal virulence factors. This review covers three categories of 23 defined mycoplasmal virulence effectors: 1) specific macromolecules including the 24 superantigen MAM, the ADP-ribosylating CARDS toxin, sialidase, cytotoxic nucleases, cell- 25 activating diacylated lipopeptides, and phosphocholine-containing glycoglycerolipids; 2) 26 the small molecule effectors hydrogen peroxide, hydrogen sulfide, and ammonia; and 3) 27 several putative mycoplasmal orthologs of virulence effectors documented in other 28 bacteria.