Interface Analysis of Small GTP Binding Protein Complexes Suggests Preferred Membrane Orientations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anti-Rab11 Antibody (ARG41900)
Product datasheet [email protected] ARG41900 Package: 100 μg anti-Rab11 antibody Store at: -20°C Summary Product Description Goat Polyclonal antibody recognizes Rab11 Tested Reactivity Hu, Ms, Rat, Dog, Mk Tested Application IHC-Fr, IHC-P, WB Host Goat Clonality Polyclonal Isotype IgG Target Name Rab11 Antigen Species Mouse Immunogen Purified recombinant peptides within aa. 110 to the C-terminus of Mouse Rab11a, Rab11b and Rab11c (Rab25). Conjugation Un-conjugated Alternate Names RAB11A: Rab-11; Ras-related protein Rab-11A; YL8 RAB11B: GTP-binding protein YPT3; H-YPT3; Ras-related protein Rab-11B RAB25: RAB11C; CATX-8; Ras-related protein Rab-25 Application Instructions Application table Application Dilution IHC-Fr 1:100 - 1:400 IHC-P 1:100 - 1:400 WB 1:250 - 1:2000 Application Note IHC-P: Antigen Retrieval: Heat mediation was recommended. * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Hepa cell lysate Calculated Mw 24 kDa Observed Size ~ 26 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer PBS, 0.05% Sodium azide and 20% Glycerol. Preservative 0.05% Sodium azide www.arigobio.com 1/3 Stabilizer 20% Glycerol Concentration 3 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. -
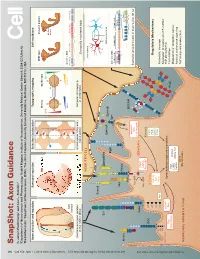
Snapshot: Axon Guidance Pasterkamp R
494 1 Cell Cell ??? SnapShot: Axon Guidance 153 SnapShot: XXXXXXXXXXXXXXXXXXXXXXXXXX 1 2 , ??MONTH?? ??DATE??, 200? ©200? Elsevier Inc. 200?©200? ElsevierInc. , ??MONTH?? ??DATE??, DOI R. Jeroen Pasterkamp and Alex L. Kolodkin , April11, 2013©2013Elsevier Inc. DOI http://dx.doi.org/10.1016/j.cell.2013.03.031 AUTHOR XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 1 AFFILIATIONDepartment of XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX Neuroscience and Pharmacology, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, 3584 CG Utrecht, The Netherlands; 2Department of Neuroscience, HHMI, The Johns Hopkins University School of Medicine, Baltimore, MD 21212, USA Axon attraction and repulsion Surround repulsion Selective fasciculation Topographic mapping Self-avoidance Wild-type Dscam1 mutant Sema3A A Retina SC/Tectum EphB EphrinB P D D Mutant T A neuron N P A V V Genomic DNA P EphA EphrinA Slit Exon 4 (12) Exon 6 (48) Exon 9 (33) Exon 17 (2) Netrin Commissural axon guidance Surround repulsion of peripheral Grasshopper CNS axon Retinotectal mapping at the CNS midline nerves in vertebrates fasciculation in vertebrates Drosophila mushroom body XXXXXXXXX NEURITE/CELL Isoneuronal Sema3 Slit Sema1/4-6 Heteroneuronal EphrinA EphrinB FasII Eph Genomic DNA Pcdh-α (14) Pcdh-β (22) Pcdh-γ (22) Variable Con Variable Con Nrp Plexin ** *** Netrin LAMELLIPODIA Con DSCAM ephexin Starburst amacrine cells in mammalian retina Ras-GTP Vav See online version for legend and references. α-chimaerin GEFs/GAPs Robo FARP Ras-GDP LARG RhoGEF Kinases DCC cc0 GTPases PKA cc1 FAK Regulatory Mechanisms See online versionfor??????. Cdc42 GSK3 cc2 Rac PI3K P1 Rho P2 cc3 Abl Proteolytic cleavage P3 Regulation of expression (TF, miRNA, FILOPODIA srGAP Cytoskeleton regulatory proteins multiple isoforms) Sos Trio Pcdh Cis inhibition DOCK180 PAK ROCK Modulation of receptors’ output LIMK Myosin-II Colin Forward and reverse signaling Actin Trafcking and endocytosis NEURONAL GROWTH CONE Microtubules SnapShot: Axon Guidance R. -

Regulation of the Mammalian Target of Rapamycin Complex 2 (Mtorc2)
Regulation of the Mammalian Target Of Rapamycin Complex 2 (mTORC2) Inauguraldissertation Zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Klaus-Dieter Molle aus Heilbronn, Deutschland Basel, 2006 Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät Auf Antrag von Prof. Michael N. Hall und Prof. Markus Affolter. Basel, den 21.11.2006 Prof. Hans-Peter Hauri Dekan Summary The growth controlling mammalian Target of Rapamycin (mTOR) is a conserved Ser/Thr kinase found in two structurally and functionally distinct complexes, mTORC1 and mTORC2. The tumor suppressor TSC1-TSC2 complex inhibits mTORC1 by acting on the small GTPase Rheb, but the role of TSC1-TSC2 and Rheb in the regulation of mTORC2 is unclear. Here we examined the role of TSC1-TSC2 in the regulation of mTORC2 in human embryonic kidney 293 cells. Induced knockdown of TSC1 and TSC2 (TSC1/2) stimulated mTORC2-dependent actin cytoskeleton organization and Paxillin phosphorylation. Furthermore, TSC1/2 siRNA increased mTORC2-dependent Ser473 phosphorylation of plasma membrane bound, myristoylated Akt/PKB. This suggests that loss of Akt/PKB Ser473 phosphorylation in TSC mutant cells, as reported previously, is due to inhibition of Akt/PKB localization rather than inhibition of mTORC2 activity. Amino acids and overexpression of Rheb failed to stimulate mTORC2 signaling. Thus, TSC1-TSC2 also inhibits mTORC2, but possibly independently of Rheb. Our results suggest that mTORC2 hyperactivation may contribute to the pathophysiology of diseases such as cancer and Tuberous Sclerosis Complex. i Acknowledgement During my PhD studies in the Biozentrum I received a lot of support from many people around me who I mention here to express my gratefulness. -

The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination
The Journal of Neuroscience, August 31, 2011 • 31(35):12579–12592 • 12579 Cellular/Molecular The Atypical Guanine-Nucleotide Exchange Factor, Dock7, Negatively Regulates Schwann Cell Differentiation and Myelination Junji Yamauchi,1,3,5 Yuki Miyamoto,1 Hajime Hamasaki,1,3 Atsushi Sanbe,1 Shinji Kusakawa,1 Akane Nakamura,2 Hideki Tsumura,2 Masahiro Maeda,4 Noriko Nemoto,6 Katsumasa Kawahara,5 Tomohiro Torii,1 and Akito Tanoue1 1Department of Pharmacology and 2Laboratory Animal Resource Facility, National Research Institute for Child Health and Development, Setagaya, Tokyo 157-8535, Japan, 3Department of Biological Sciences, Tokyo Institute of Technology, Midori, Yokohama 226-8501, Japan, 4IBL, Ltd., Fujioka, Gumma 375-0005, Japan, and 5Department of Physiology and 6Bioimaging Research Center, Kitasato University School of Medicine, Sagamihara, Kanagawa 252-0374, Japan In development of the peripheral nervous system, Schwann cells proliferate, migrate, and ultimately differentiate to form myelin sheath. In all of the myelination stages, Schwann cells continuously undergo morphological changes; however, little is known about their underlying molecular mechanisms. We previously cloned the dock7 gene encoding the atypical Rho family guanine-nucleotide exchange factor (GEF) and reported the positive role of Dock7, the target Rho GTPases Rac/Cdc42, and the downstream c-Jun N-terminal kinase in Schwann cell migration (Yamauchi et al., 2008). We investigated the role of Dock7 in Schwann cell differentiation and myelination. Knockdown of Dock7 by the specific small interfering (si)RNA in primary Schwann cells promotes dibutyryl cAMP-induced morpholog- ical differentiation, indicating the negative role of Dock7 in Schwann cell differentiation. It also results in a shorter duration of activation of Rac/Cdc42 and JNK, which is the negative regulator of myelination, and the earlier activation of Rho and Rho-kinase, which is the positive regulator of myelination. -

Figure S1. DMD Module Network. the Network Is Formed by 260 Genes from Disgenet and 1101 Interactions from STRING. Red Nodes Are the Five Seed Candidate Genes
Figure S1. DMD module network. The network is formed by 260 genes from DisGeNET and 1101 interactions from STRING. Red nodes are the five seed candidate genes. Figure S2. DMD module network is more connected than a random module of the same size. It is shown the distribution of the largest connected component of 10.000 random modules of the same size of the DMD module network. The green line (x=260) represents the DMD largest connected component, obtaining a z-score=8.9. Figure S3. Shared genes between BMD and DMD signature. A) A meta-analysis of three microarray datasets (GSE3307, GSE13608 and GSE109178) was performed for the identification of differentially expressed genes (DEGs) in BMD muscle biopsies as compared to normal muscle biopsies. Briefly, the GSE13608 dataset included 6 samples of skeletal muscle biopsy from healthy people and 5 samples from BMD patients. Biopsies were taken from either biceps brachii, triceps brachii or deltoid. The GSE3307 dataset included 17 samples of skeletal muscle biopsy from healthy people and 10 samples from BMD patients. The GSE109178 dataset included 14 samples of controls and 11 samples from BMD patients. For both GSE3307 and GSE10917 datasets, biopsies were taken at the time of diagnosis and from the vastus lateralis. For the meta-analysis of GSE13608, GSE3307 and GSE109178, a random effects model of effect size measure was used to integrate gene expression patterns from the two datasets. Genes with an adjusted p value (FDR) < 0.05 and an │effect size│>2 were identified as DEGs and selected for further analysis. A significant number of DEGs (p<0.001) were in common with the DMD signature genes (blue nodes), as determined by a hypergeometric test assessing the significance of the overlap between the BMD DEGs and the number of DMD signature genes B) MCODE analysis of the overlapping genes between BMD DEGs and DMD signature genes. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

LPS Induces GFAT2 Expression to Promote O-Glcnacylation
1 LPS induces GFAT2 expression to promote O-GlcNAcylation and 2 attenuate inflammation in macrophages 3 4 5 Running title: GFAT2 is a FoxO1-dependent LPS-inducible gene 6 7 Hasanain AL-MUKH, Léa BAUDOIN, Abdelouhab BOUABOUD, José-Luis SANCHEZ- 8 SALGADO, Nabih MARAQA, Mostafa KHAIR, Patrick PAGESY, Georges BISMUTH, 9 Florence NIEDERGANG and Tarik ISSAD 10 11 Université de Paris, Institut Cochin, CNRS, INSERM, F-75014 Paris, France 12 Address correspondence to: Tarik Issad, Institut Cochin, Department of Endocrinology, 13 Metabolism and Diabetes, 24 rue du Faubourg Saint-Jacques, 75014 Paris FRANCE. Tel : + 33 1 14 44 41 25 67; E-mail: [email protected] 15 16 1 1 Abstract 2 O-GlcNAc glycosylation is a reversible post-translational modification that regulates the 3 activity of intracellular proteins according to glucose availability and its metabolism through 4 the hexosamine biosynthesis pathway (HBP). This modification has been involved in the 5 regulation of various immune cell types, including macrophages. However, little is known 6 concerning the mechanisms that regulate protein O-GlcNAcylation level in these cells. In the 7 present work, we demonstrate that LPS treatment induces a marked increase in protein O- 8 GlcNAcylation in RAW264.7 cells, bone-marrow-derived and peritoneal mouse 9 macrophages, as well as human monocyte-derived macrophages. Targeted deletion of OGT in 10 macrophages resulted in an increased effect of LPS on NOS2 expression and cytokine 11 production, suggesting that O-GlcNAcylation may restrain inflammatory processes induced 12 by LPS. The effect of LPS on protein O-GlcNAcylation in macrophages was associated with 13 an increased expression and activity of glutamine fructose 6-phosphate amido-transferase 14 (GFAT), the enzyme that catalyzes the rate-limiting step of the HBP. -

Cell, Volume 139 Supplemental Data Profiling the Human Protein-DNA
Cell, Volume 139 Supplemental Data Profiling the Human Protein-DNA Interactome Reveals ERK2 as a Transcriptional Repressor of Interferon Signaling Shaohui Hu, Zhi Xie, Akishi Onishi, Xueping Yu, Lizhi Jiang, Jimmy Lin, Hee-sool Rho, Crystal Woodard, Hong Wang, Jun-Seop Jeong, Shunyou Long, Xiaofei He, Herschel Wade, Seth Blackshaw, Jiang Qian, and Heng Zhu Supplemental Experimental Procedures Identifying Tissue-specific Motifs We developed a program to identify tissue-specific motifs. We first defined sets of tissue-specific or tissue-enriched genes by examining their gene expression profiles across multiple tissues (Yu et al., 2006). We then calculated the most over-represented single motifs (8-mers, including a wide character) in the promoters of each set of tissue-specific genes. The program then enumerated all possible combinations of the top n motifs (e.g. n = 100). For each motif pair, the program recorded the occurrence of the motif pair in the promoter sequences. We then calculated the significance score for each motif pair, which was defined as the negative logarithm of the p value, -log(p). The motif pairs with scores above a specified threshold were considered putative TF binding motif pairs in the promoter sequences. With these predicted motif pairs, we could calculate a number of partners for each motif and select a certain number of top non-redundant motifs to be tested in the protein chip experiments. Both the p values for a single motif and those for a motif pair were calculated using hypergeometric distribution. Here, we use a motif pair as an example to show the ij, procedure. -

A Role for N-Myristoylation in Protein Targeting
A Role for N-Myristoylation in Protein Targeting: NADH-Cytochrome b5 Reductase Requires Myristic Acid for Association with Outer Mitochondrial But Not ER Membranes Nica Borgese,** Diego Aggujaro,* Paola Carrera, ~ Grazia Pietrini,* and Monique Bassetti* *Consiglio Nazionale deUe Ricerche Cellular and Molecular Pharmacology Center, Department of Pharmacology, University of Milan, 20129 Milan, Italy; ~Faculty of Pharmacy, University of Reggio Calabria, 88021 Roccelletta di Borgia, Catanzaro, Italy; and ~Scientific Institute "San Raffaele," 20132 Milan, Italy Abstract. N-myristoylation is a cotranslational modifi- investigated by immunofluorescence, immuno-EM, and Downloaded from http://rupress.org/jcb/article-pdf/135/6/1501/1267948/1501.pdf by guest on 27 September 2021 cation involved in protein-protein interactions as well cell fractionation. By all three techniques, the wt pro- as in anchoring polypeptides to phospholipid bilayers; tein localized to ER and mitochondria, while the non- however, its role in targeting proteins to specific subcel- myristoylated mutant was found only on ER mem- lular compartments has not been clearly defined. The branes. Pulse-chase experiments indicated that this mammalian myristoylated flavoenzyme NADH-cyto- altered steady state distribution was due to the mu- chrome b 5 reductase is integrated into ER and mito- tant's inability to target to mitochondria, and not to its chondrial outer membranes via an anchor containing a enhanced instability in that location. Both wt and mu- stretch of 14 uncharged amino acids downstream to the tant reductase were resistant to Na2CO 3 extraction and NH2-terminal myristoylated glycine. Since previous partitioned into the detergent phase after treatment of studies suggested that the anchoring function could be a membrane fraction with Triton X-114, demonstrating adequately carried out by the 14 uncharged residues, that myristic acid is not required for tight anchoring of we investigated a possible role for myristic acid in re- reductase to membranes. -

RNF11 at the Crossroads of Protein Ubiquitination
biomolecules Review RNF11 at the Crossroads of Protein Ubiquitination Anna Mattioni, Luisa Castagnoli and Elena Santonico * Department of Biology, University of Rome Tor Vergata, Via della ricerca scientifica, 00133 Rome, Italy; [email protected] (A.M.); [email protected] (L.C.) * Correspondence: [email protected] Received: 29 September 2020; Accepted: 8 November 2020; Published: 11 November 2020 Abstract: RNF11 (Ring Finger Protein 11) is a 154 amino-acid long protein that contains a RING-H2 domain, whose sequence has remained substantially unchanged throughout vertebrate evolution. RNF11 has drawn attention as a modulator of protein degradation by HECT E3 ligases. Indeed, the large number of substrates that are regulated by HECT ligases, such as ITCH, SMURF1/2, WWP1/2, and NEDD4, and their role in turning off the signaling by ubiquitin-mediated degradation, candidates RNF11 as the master regulator of a plethora of signaling pathways. Starting from the analysis of the primary sequence motifs and from the list of RNF11 protein partners, we summarize the evidence implicating RNF11 as an important player in modulating ubiquitin-regulated processes that are involved in transforming growth factor beta (TGF-β), nuclear factor-κB (NF-κB), and Epidermal Growth Factor (EGF) signaling pathways. This connection appears to be particularly significant, since RNF11 is overexpressed in several tumors, even though its role as tumor growth inhibitor or promoter is still controversial. The review highlights the different facets and peculiarities of this unconventional small RING-E3 ligase and its implication in tumorigenesis, invasion, neuroinflammation, and cancer metastasis. Keywords: Ring Finger Protein 11; HECT ligases; ubiquitination 1. -

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between