Features of Lexical Richness in Children's Books: Comparisons with Child-Directed Speech
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Applied Category Theory for Genomics – an Initiative
Applied Category Theory for Genomics { An Initiative Yanying Wu1,2 1Centre for Neural Circuits and Behaviour, University of Oxford, UK 2Department of Physiology, Anatomy and Genetics, University of Oxford, UK 06 Sept, 2020 Abstract The ultimate secret of all lives on earth is hidden in their genomes { a totality of DNA sequences. We currently know the whole genome sequence of many organisms, while our understanding of the genome architecture on a systematic level remains rudimentary. Applied category theory opens a promising way to integrate the humongous amount of heterogeneous informations in genomics, to advance our knowledge regarding genome organization, and to provide us with a deep and holistic view of our own genomes. In this work we explain why applied category theory carries such a hope, and we move on to show how it could actually do so, albeit in baby steps. The manuscript intends to be readable to both mathematicians and biologists, therefore no prior knowledge is required from either side. arXiv:2009.02822v1 [q-bio.GN] 6 Sep 2020 1 Introduction DNA, the genetic material of all living beings on this planet, holds the secret of life. The complete set of DNA sequences in an organism constitutes its genome { the blueprint and instruction manual of that organism, be it a human or fly [1]. Therefore, genomics, which studies the contents and meaning of genomes, has been standing in the central stage of scientific research since its birth. The twentieth century witnessed three milestones of genomics research [1]. It began with the discovery of Mendel's laws of inheritance [2], sparked a climax in the middle with the reveal of DNA double helix structure [3], and ended with the accomplishment of a first draft of complete human genome sequences [4]. -

Functional Effects Detailed Research Plan
GeCIP Detailed Research Plan Form Background The Genomics England Clinical Interpretation Partnership (GeCIP) brings together researchers, clinicians and trainees from both academia and the NHS to analyse, refine and make new discoveries from the data from the 100,000 Genomes Project. The aims of the partnerships are: 1. To optimise: • clinical data and sample collection • clinical reporting • data validation and interpretation. 2. To improve understanding of the implications of genomic findings and improve the accuracy and reliability of information fed back to patients. To add to knowledge of the genetic basis of disease. 3. To provide a sustainable thriving training environment. The initial wave of GeCIP domains was announced in June 2015 following a first round of applications in January 2015. On the 18th June 2015 we invited the inaugurated GeCIP domains to develop more detailed research plans working closely with Genomics England. These will be used to ensure that the plans are complimentary and add real value across the GeCIP portfolio and address the aims and objectives of the 100,000 Genomes Project. They will be shared with the MRC, Wellcome Trust, NIHR and Cancer Research UK as existing members of the GeCIP Board to give advance warning and manage funding requests to maximise the funds available to each domain. However, formal applications will then be required to be submitted to individual funders. They will allow Genomics England to plan shared core analyses and the required research and computing infrastructure to support the proposed research. They will also form the basis of assessment by the Project’s Access Review Committee, to permit access to data. -

Hmms Representing All Proteins of Known Structure. SCOP Sequence Searches, Alignments and Genome Assignments Julian Gough* and Cyrus Chothia
268–272 Nucleic Acids Research, 2002, Vol. 30, No. 1 © 2002 Oxford University Press SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments Julian Gough* and Cyrus Chothia MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, UK Received September 28, 2001; Revised and Accepted October 30, 2001 ABSTRACT (currently 59) covering approximately half of the The SUPERFAMILY database contains a library of soluble protein domains. The assignments, super- family breakdown and statistics on them are available hidden Markov models representing all proteins of from the server. The database is currently used by known structure. The database is based on the SCOP this group and others for genome annotation, structural ‘superfamily’ level of protein domain classification genomics, gene prediction and domain-based genomic which groups together the most distantly related studies. proteins which have a common evolutionary ancestor. There is a public server at http://supfam.org which provides three services: sequence searching, INTRODUCTION multiple alignments to sequences of known structure, The SUPERFAMILY database is based on the SCOP (1) and structural assignments to all complete genomes. classification of protein domains. SCOP is a structural domain- Given an amino acid or nucleotide query sequence based heirarchical classification with several levels including the server will return the domain architecture and the ‘superfamily’ level. Proteins grouped together at the super- SCOP classification. The server produces alignments family level are defined as having structural, functional and sequence evidence for a common evolutionary ancestor. It is at of the query sequences with sequences of known this level, as the name suggests, that SUPERFAMILY operates structure, and includes multiple alignments of because it is the level with the most distantly related protein genome and PDB sequences. -

AUTOPHY by Deepika Prasad a Thesis
ANALYZING MARKER GENE DIVERSITY USING AN AUTOMATED PHYLOGENETIC TOOL: AUTOPHY by Deepika Prasad A thesis submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the degree of Master of Science in Bioinformatics and Computational Biology Spring 2017 © 2017 Deepika Prasad All Rights Reserved ANALYZING MARKER GENE DIVERSITY USING AN AUTOMATED PHYLOGENETIC TOOL: AUTOPHY by Deepika Prasad Approved: __________________________________________________________ Shawn Polson, Ph.D. Professor in charge of thesis on behalf of the Advisory Committee Approved: __________________________________________________________ Kathleen F.McCoy, Ph.D. Chair of the Department of Computer and Information Sciences Approved: __________________________________________________________ Babatunde A. Ogunnaike, Ph.D. Dean of the College of Engineering Approved: __________________________________________________________ Ann L. Ardis, Ph.D. Senior Vice Provost for Graduate and Professional Education ACKNOWLEDGMENTS I want to express my sincerest gratitude to Professor Shawn Polson for his patience, continuous support, and enthusiasm. I could not have imagined having a better mentor to guide me through the process of my Master’s thesis. I would like to thank my committee members, Professor Eric Wommack and Honzhan Huang for their insightful comments and guidance in the thesis. I would also like to express my gratitude to Barbra Ferrell for asking important questions throughout my thesis research and helping me with the writing process. I would like to thank my friend Sagar Doshi, and lab mates Daniel Nasko, and Prasanna Joglekar, for helping me out with data, presentations, and giving important advice, whenever necessary. Last but not the least, I would like to thank my family, for being my pillars of strength. -

Booktrust Laureate List
Celebrate the 20th anniversary of the Waterstones Children’s Laureate with these Laureate books with a travelling theme… Suggested by Emily Drabble, BookTrust’s head of Children’s Book Promotions and Prizes Quentin Blake The Green Ship (Penguin) When two children journey over the wall, they discover a ship made out of trees and go on an imaginative voyage Mrs Armitage of Wheels (Penguin) Mrs Armitage sets off on a quiet cycle with her dog Breakspear but can’t help thinking of ways to improve her bike with disastrous consequences! Anne Fine Mrs Doubtfire (Puffin) lots of travelling from house to house for the split family in this masterpiece, and of course the journey the impractical out-of-work actor Dad Daniel goes on in this book is huge. Killer Cat Runs Away (Puffin) Tuffy the cat doesn’t feel at home anymore after he gets In big trouble from breaking the TV and eating Tinkerbell’s special kitten, so Tuffy decides to make a break for it and run away! Jacqueline Wilson Cliffhanger (Penguin, illustrated by Nick Sharratt) an adventure holiday sounds like fun, but Tim proves to be hopeless at sports of any kind. Can he survive the horrors of the week? Buried Alive (Penguin, illustrated by Nick Sharratt) Tim gets to bring his friend Biscuits on holiday with his family. But some local bully boys try and disrupt the fun. Michael Morpurgo Running Wild (Harper Collins) Will’s holiday to Indonesia with his mum turns to disaster as the Boxing Day Tsunami hits – and his escape on the back of Oona the elephant begins a truly epic adventure. -
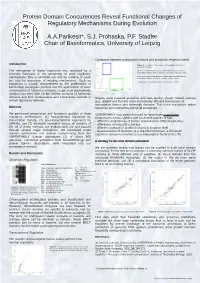
Protein Domain Coocurences Reveal Functional Changes of Regulatory Mechanisms During Evolution
Protein Domain Coocurences Reveal Functional Changes of Regulatory Mechanisms During Evolution A.A.Parikesit*, S.J. Prohaska, P.F. Stadler Chair of Bioinformatics, University of Leipzig Correlation between transcription factors and chromatin related proteins Introduction Figure 3 : Correlation of the number of transcription factors and chromatin domains. The emergence of higher organisms was facilitated by a Blue box. Species with few chromatin-related domains but many dramatic increases in the complexity of gene regulatory transcription factors: human, kangaroo rat, mouse, opossum, fugu. Green box: species with many chromatin-related domains but few mechanisms. This is achieved not only by addition of novel transcription factors: seq squirt, medaka, dolphin, yeast but also by expansion of existing mechanisms. Such an There are no organisms (known) that have a lot of both. expansion is usually characterized by the proliferation of functionally paralogous proteins and the appearance of novel combinations of functional domains. Large scale phylogenetic analysis can shed light on the relative amounts of functional domains and their combinations and interactions involved in Results show massive problems with data quality: closely related species certain regulatory networks. (e.g, dolphin and human) show dramatically different distributions of transcription factors and chromatin domains. This is not reasonable within Methods mammals and contradicts biological knowledge. We performed comparative and functional analysis of three SUPERFAMILY thus cannot be used for large-scale quantitative regulatory mechanisms: (1) transcriptional regulation by comparisons across species due to several sources of bias: transcription factors, (2) post-transcriptional regulation by - different completeness of protein annotation for different genomes miRNAs, and (3) chromatin regulation across all domains of - differences in transcript coverage life. -

33Rd IBBY International Congress, London
35 AUTUMN 2012 33rd IBBY International Congress, London CONTENTS EDITORIAL 2 Second Day of the Congress, Saturday 18 Impressions of the 33rd IBBY International Ellen Ainsworth Congress: ‘Crossing Boundaries, Translations Third and Final Day of the Congress, Sunday 19 and Migrations’ 3 Jaq Delany Darja Mazi-Leskovar, Slovenia 3 Clive Barnes, UK 4 Post-Congress Excursion, Tuesday: Discovering the Real ‘Green Knowe’ and ‘Midnight Valerie Coghlan, Ireland 5 Garden’ 20 Petros Panaou, Cyprus 6 Ellen Ainsworth Alice Curry, UK 6 Strange Migrations 22 Niklas Bengtsson, Finland 7 Shaun Tan Pam Dix, UK 8 Pat Pinsent, UK 8 REVIEWS 32 Rebecca R. Butler, UK 9 REPORTS 43 Swapna Dutta, India 10 AWARDS 44 Judith Philo, UK 11 FORTHCOMING EXHIBITIONS AND EVENTS 55 Ferelith Hordon, UK 12 NEWS 58 Susan Bailes, UK 13 IBBY NEWS 61 First Day of the Congress, Friday 15 Alexandra Strick 15 Erica Gillingham 17 Logo for the 33rd IBBY International Congress held at Imperial College London. Designed by former Children’s Laureate (2009–2011) Anthony Browne. EDITORIAL ‘ There is in London all that life can afford’ illustrator, to put into words the feelings so well (Samuel Johnson, 1777, quoted by Boswell) conveyed in his own pictures, meant that he Certainly Dr Johnson’s words could well be applied encapsulated the experience of so many of the to London in summer 2012, with the Jubilee, the children with whom IBBY is concerned. Olympics and Paralympics, and, more pertinent to The congress isn’t the only thing happening this IBBYLink, the 33rd IBBY International Congress. year – details of the annual November Many people have spoken about how heartening it IBBY/NCRCL MA conference at Roehampton are was to see so many people from different parts of given on p.61 and on the back cover. -

Renaissance SUPERFAMILY in the Decade Since Its Launch, a Repository of Information About Proteins in Genomes Has Developed Into a Primary Reference
Renaissance SUPERFAMILY In the decade since its launch, a repository of information about proteins in genomes has developed into a primary reference. Dr Julian Gough, its creator, describes current work enhancing the scope, content and functionality of this key service Could you begin by outlining the reasons for How do HMMs feed into the service? creating SUPERFAMILY? HMMs are profi les which represent multiple SUPERFAMILY was originally created to better sequence alignments of homologous proteins understand molecular evolution, initially by in a rigorous statistical framework. They enabling comparison of the repertoire of proteins can be used to classify sequences based on and domains across the genomes of different homology and to create sequence alignments. species. The starting point was the Structural We use sequences of domains of known Classifi cation of Proteins (SCOP), and the most structure via iterative search procedures on basic purpose of SUPERFAMILY is to detect and large background sequence databases to classify these domains with known structural build alignments and, subsequently, models representatives in the protein sequences of representing domains of known structure at genomes. There are tens of thousands of known the superfamily level. These models are then protein structures, each the result of a costly searched against genome sequences to detect three-dimensional atomic resolution structure and classify the structural domains. determination by experiment, usually X-ray crystallography or nuclear magnetic resonance. -

An Expanded Evaluation of Protein Function Prediction Methods Shows an Improvement in Accuracy
The University of Southern Mississippi The Aquila Digital Community Faculty Publications 9-7-2016 An Expanded Evaluation of Protein Function Prediction Methods Shows an Improvement In Accuracy Yuxiang Jiang Indiana University TFollowal Ronnen this and Or onadditional works at: https://aquila.usm.edu/fac_pubs Buck P arInstitutet of the forGenomics Research Commons On Agin Wyatt T. Clark RecommendedYale University Citation AsmaJiang, YR.., OrBankapuron, T. R., Clark, W. T., Bankapur, A. R., D'Andrea, D., Lepore, R., Funk, C. S., Kahanda, I., Verspoor, K. M., Ben-Hur, A., Koo, D. E., Penfold-Brown, D., Shasha, D., Youngs, N., Bonneau, R., Lin, A., Sahraeian, S. Miami University M., Martelli, P. L., Profiti, G., Casadio, R., Cao, R., Zhong, Z., Cheng, J., Altenhoff, A., Skunca, N., Dessimoz, DanielC., Dogan, D'Andr T., Hakala,ea K., Kaewphan, S., Mehryar, F., Salakoski, T., Ginter, F., Fang, H., Smithers, B., Oates, M., UnivGough,ersity J., ofTör Romeönen, P., Koskinen, P., Holm, L., Chen, C., Hsu, W., Bryson, K., Cozzetto, D., Minneci, F., Jones, D. T., Chapan, S., BKC, D., Khan, I. K., Kihara, D., Ofer, D., Rappoport, N., Stern, A., Cibrian-Uhalte, E., Denny, P., Foulger, R. E., Hieta, R., Legge, D., Lovering, R. C., Magrane, M., Melidoni, A. N., Mutowo-Meullenet, P., SeePichler next, K., page Shypitsyna, for additional A., Li, B.,authors Zakeri, P., ElShal, S., Tranchevent, L., Das, S., Dawson, N. L., Lee, D., Lees, J. G., Stilltoe, I., Bhat, P., Nepusz, T., Romero, A. E., Sasidharan, R., Yang, H., Paccanaro, A., Gillis, J., Sedeño- Cortés, A. E., Pavlidis, P., Feng, S., Cejuela, J. -

Simulation and Graph Mining Tools for Improving Gene Mapping Efficiency
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Helsingin yliopiston digitaalinen arkisto DEPARTMENT OF COMPUTER SCIENCE SERIES OF PUBLICATIONS A REPORT A-2011-3 Simulation and graph mining tools for improving gene mapping efficiency Petteri Hintsanen To be presented, with the permission of the Faculty of Science of the University of Helsinki, for public criticism in Auditorium XIV (Univer- sity Main Building, Unioninkatu 34) on 30 September 2011 at twelve o’clock noon. UNIVERSITY OF HELSINKI FINLAND Supervisor Hannu Toivonen, University of Helsinki, Finland Pre-examiners Tapio Salakoski, University of Turku, Finland Janne Nikkila,¨ University of Helsinki, Finland Opponent Michael Berthold, Universitat¨ Konstanz, Germany Custos Hannu Toivonen, University of Helsinki, Finland Contact information Department of Computer Science P.O. Box 68 (Gustaf Hallstr¨ omin¨ katu 2b) FI-00014 University of Helsinki Finland Email address: [email protected].fi URL: http://www.cs.Helsinki.fi/ Telephone: +358 9 1911, telefax: +358 9 191 51120 Copyright c 2011 Petteri Hintsanen ISSN 1238-8645 ISBN 978-952-10-7139-3 (paperback) ISBN 978-952-10-7140-9 (PDF) Computing Reviews (1998) Classification: G.2.2, G.3, H.2.5, H.2.8, J.3 Helsinki 2011 Helsinki University Print Simulation and graph mining tools for improving gene mapping efficiency Petteri Hintsanen Department of Computer Science P.O. Box 68, FI-00014 University of Helsinki, Finland petteri.hintsanen@iki.fi http://iki.fi/petterih PhD Thesis, Series of Publications A, Report A-2011-3 Helsinki, September 2011, 136 pages ISSN 1238-8645 ISBN 978-952-10-7139-3 (paperback) ISBN 978-952-10-7140-9 (PDF) Abstract Gene mapping is a systematic search for genes that affect observable character- istics of an organism. -

Specialized Hidden Markov Model Databases for Microbial Genomics
Comparative and Functional Genomics Comp Funct Genom 2003; 4: 250–254. Published online 1 April 2003 in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/cfg.280 Conference Review Specialized hidden Markov model databases for microbial genomics Martin Gollery* University of Nevada, Reno, 1664 N. Virginia Street, Reno, NV 89557-0014, USA *Correspondence to: Abstract Martin Gollery, University of Nevada, Reno, 1664 N. Virginia As hidden Markov models (HMMs) become increasingly more important in the Street, Reno, NV analysis of biological sequences, so too have databases of HMMs expanded in size, 89557-0014, USA. number and importance. While the standard paradigm a short while ago was the E-mail: [email protected] analysis of one or a few sequences at a time, it has now become standard procedure to submit an entire microbial genome. In the future, it will be common to submit large groups of completed genomes to run simultaneously against a dozen public databases and any number of internally developed targets. This paper looks at some of the readily available HMM (or HMM-like) algorithms and several publicly available Received: 27 January 2003 HMM databases, and outlines methods by which the reader may develop custom Revised: 5 February 2003 HMM targets. Copyright 2003 John Wiley & Sons, Ltd. Accepted: 6 February 2003 Keywords: HMM; Pfam; InterPro; SuperFamily; TLfam; COG; TIGRfams Introduction will be a true homologue. As a result, HMMs have become very popular in the field of bioinformatics Over the last few years, hidden Markov models and a number of HMM databases have been (HMMs) have become one of the pre-eminent developed. -

Annual Review 2011-2012 Year at a Glance 1
Annual Review 2011-2012 Year at a glance 1 In August 2011, a team led by Professor Andrew Baker was awarded £3.9m for research into preventing a common failure of heart bypass grafts. Scientists, cardiologists and surgeons from the University and the Golden Jubilee National Principal’s welcome Hospital will undertake the first ever trial of a new gene therapy – 15 years in the making – in human patients. Welcome to the University of Glasgow’s Annual Review, which is a brief guide to what has been a very successful year, covering the period August 2011 to July 2012. September I am pleased to report on the continuing financial stability of the University, and the operating surpluses we have secured to invest in our teaching, infrastructure and research. The breadth of our expertise is a key strength and one you’ll see reflected in the diverse range of stories we have to tell. In much of our work we make progress through collaboration, both within and beyond our community. This year, Glasgow has become more connected to the world than ever before. The University’s museum and art gallery was reopened after a We are proud to have signed partnership agreements with, among others, University of two-year period of refurbishment. Calcutta, Singapore Institute of Technology, the Smithsonian Institution and George Mason University in order to enable the exchange of staff, students and ideas. The Hunterian is Scotland’s oldest public museum. The magnificent space now hosts a new gallery featuring a unique collection of Roman artefacts recovered from the Antonine Wall.