11 Matrix Arithmetic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Handbook of Double Stars, with a Catalogue of Twelve Hundred
The original of this bool< is in the Cornell University Library. There are no known copyright restrictions in the United States on the use of the text. http://www.archive.org/details/cu31924064295326 3 1924 064 295 326 Production Note Cornell University Library pro- duced this volume to replace the irreparably deteriorated original. It was scanned using Xerox soft- ware and equipment at 600 dots per inch resolution and com- pressed prior to storage using CCITT Group 4 compression. The digital data were used to create Cornell's replacement volume on paper that meets the ANSI Stand- ard Z39. 48-1984. The production of this volume was supported in part by the Commission on Pres- ervation and Access and the Xerox Corporation. Digital file copy- right by Cornell University Library 1991. HANDBOOK DOUBLE STARS. <-v6f'. — A HANDBOOK OF DOUBLE STARS, WITH A CATALOGUE OF TWELVE HUNDRED DOUBLE STARS AND EXTENSIVE LISTS OF MEASURES. With additional Notes bringing the Measures up to 1879, FOR THE USE OF AMATEURS. EDWD. CROSSLEY, F.R.A.S.; JOSEPH GLEDHILL, F.R.A.S., AND^^iMES Mt^'^I^SON, M.A., F.R.A.S. "The subject has already proved so extensive, and still ptomises so rich a harvest to those who are inclined to be diligent in the pursuit, that I cannot help inviting every lover of astronomy to join with me in observations that must inevitably lead to new discoveries." Sir Wm. Herschel. *' Stellae fixac, quae in ccelo conspiciuntur, sunt aut soles simplices, qualis sol noster, aut systemata ex binis vel interdum pluribus solibus peculiari nexu physico inter se junccis composita. -

The Minor Planet Bulletin 44 (2017) 142
THE MINOR PLANET BULLETIN OF THE MINOR PLANETS SECTION OF THE BULLETIN ASSOCIATION OF LUNAR AND PLANETARY OBSERVERS VOLUME 44, NUMBER 2, A.D. 2017 APRIL-JUNE 87. 319 LEONA AND 341 CALIFORNIA – Lightcurves from all sessions are then composited with no TWO VERY SLOWLY ROTATING ASTEROIDS adjustment of instrumental magnitudes. A search should be made for possible tumbling behavior. This is revealed whenever Frederick Pilcher successive rotational cycles show significant variation, and Organ Mesa Observatory (G50) quantified with simultaneous 2 period software. In addition, it is 4438 Organ Mesa Loop useful to obtain a small number of all-night sessions for each Las Cruces, NM 88011 USA object near opposition to look for possible small amplitude short [email protected] period variations. Lorenzo Franco Observations to obtain the data used in this paper were made at the Balzaretto Observatory (A81) Organ Mesa Observatory with a 0.35-meter Meade LX200 GPS Rome, ITALY Schmidt-Cassegrain (SCT) and SBIG STL-1001E CCD. Exposures were 60 seconds, unguided, with a clear filter. All Petr Pravec measurements were calibrated from CMC15 r’ values to Cousins Astronomical Institute R magnitudes for solar colored field stars. Photometric Academy of Sciences of the Czech Republic measurement is with MPO Canopus software. To reduce the Fricova 1, CZ-25165 number of points on the lightcurves and make them easier to read, Ondrejov, CZECH REPUBLIC data points on all lightcurves constructed with MPO Canopus software have been binned in sets of 3 with a maximum time (Received: 2016 Dec 20) difference of 5 minutes between points in each bin. -

Final Report Asteroid Impact Monitoring
Final Report Asteroid Impact Monitoring Environmental and Instrumentation Requirements A component of the Asteroid Impact & Deflection Assessment (AIDA) Mission By the AIM Advisory Team Dr. Patrick MICHEL (Univ. Nice, CNRS, OCA), Team Leader Dr. Jens Biele (DLR) Dr. Marco Delbo (Univ. Nice, CNRS, OCA) Dr. Martin Jutzi (Univ. Bern) Pr. Guy Libourel (Univ. Nice, CNRS, OCA) Dr. Naomi Murdoch Dr. Stephen R. Schwartz (Univ. Nice, CNRS, OCA) Dr. Stephan Ulamec (DLR) Dr. Jean-Baptiste Vincent (MPS) April 12th, 2014 Introduction In this report, we describe the knowledge gain resulting from the implementation of either the European Space Agency’s Asteroid Impact Monitoring (AIM) as a stand- alone mission or AIM with its second component, the Double Asteroid Redirection Test (DART) mission under study by the Johns Hopkins Applied Physics Laboratory with support from members of NASA centers including Goddard Space Flight Center, Johnson Space Center, and the Jet Propulsion Laboratory. We then present our analysis of the required measurements addressing the goals of the AIM mission to the binary Near-Earth Asteroid (NEA) Didymos, and for two specified payloads. The first payload is a mini thermal infrared camera (called TP1) for short and medium range characterisation. The second payload is an active seismic experiment (called TP2). We then present the environmental parameters for the AIM mission. AIM is a rendezvous mission that focuses on the monitoring aspects i.e., the capability to determine in-situ the key properties of the secondary of a binary asteroid. DART consists primarily of an artificial projectile aims to demonstrate asteroid deflection. In the framework of the full AIDA concept, AIM will also give access to the detailed conditions of the DART impact and its outcome, allowing for the first time to get a complete picture of such an event, a better interpretation of the deflection measurement and a possibility to compare with numerical modeling predictions. -

1 PRELIMINARY SPIN-SHAPE MODEL for 755 QUINTILLA Lorenzo Franco Balzaretto Observatory (A81), Rome, ITALY Lor [email protected] R
1 PRELIMINARY SPIN-SHAPE MODEL FOR Lightcurve inversion was performed using MPO LCInvert 755 QUINTILLA v.11.8.2.0 (BDW Publishing, 2016). For a description of the modeling process see LCInvert Operating Instructions Manual, Lorenzo Franco Durech et al. (2010); and references therein. Balzaretto Observatory (A81), Rome, ITALY [email protected] Figure 1 shows the PAB longitude/latitude distribution for dense/sparse data used in the lightcurve inversion process. Figure Robert K. Buchheim 2 (top panel) shows the sparse photometric data distribution Altimira Observatory (G76) (intensities vs JD) and (bottom panel) the corresponding phase 18 Altimira, Coto de Caza, CA 92679 curve (reduced magnitudes vs phase angle). Donald Pray Carbuncle Hill Observatory (I00) Greene, Rhode Island Michael Fauerbach Florida Gulf Coast University 10501 FGCU Blvd. Ft. Myers, FL33965-6565 Fabio Mortari Hypatia Observatory (L62), Rimini, ITALY Giovanni Battista Casalnuovo, Benedetto Chinaglia Filzi School Observatory (D12), Laives, ITALY Figure 1: PAB longitude and latitude distribution of the data used Giulio Scarfi for the lightcurve inversion model. Iota Scorpii Observatory (K78), La Spezia, ITALY Riccardo Papini, Fabio Salvaggio Wild Boar Remote Observatory (K49) San Casciano in Val di Pesa (FI), ITALY (Received: Revised: ) We present a preliminary shape and spin axis model for main-belt asteroid 755 Quintilla. The model was achieved with the lightcurve inversion process, using combined dense photometric data acquired from three apparitions, between 2004 to 2020 and sparse data from USNO Flagstaff. Analysis of the resulting data found a sidereal period P = 4.55204 ± 0.00001 hours and two mirrored pole solutions at (λ = 109°, β = -12°) and (λ = 288°, β = -3°) with an uncertainty of ± 20 degrees. -

Applying Machine Learning to Asteroid Classification Utilizing Spectroscopically Derived Spectrophotometry Kathleen Jacinda Mcintyre
University of North Dakota UND Scholarly Commons Theses and Dissertations Theses, Dissertations, and Senior Projects January 2019 Applying Machine Learning To Asteroid Classification Utilizing Spectroscopically Derived Spectrophotometry Kathleen Jacinda Mcintyre Follow this and additional works at: https://commons.und.edu/theses Recommended Citation Mcintyre, Kathleen Jacinda, "Applying Machine Learning To Asteroid Classification Utilizing Spectroscopically Derived Spectrophotometry" (2019). Theses and Dissertations. 2474. https://commons.und.edu/theses/2474 This Thesis is brought to you for free and open access by the Theses, Dissertations, and Senior Projects at UND Scholarly Commons. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of UND Scholarly Commons. For more information, please contact [email protected]. APPLYING MACHINE LEARNING TO ASTEROID CLASSIFICATION UTILIZING SPECTROSCOPICALLY DERIVED SPECTROPHOTOMETRY by Kathleen Jacinda McIntyre Bachelor oF Science, University oF Florida, 2011 Bachelor oF Arts, University oF Florida, 2011 A Thesis Submitted to the Graduate Faculty of the University oF North Dakota in partial fulfillment oF the reQuirements for the degree oF Master oF Science Grand Forks, North Dakota May 2019 ii PERMISSION Title ApPlying Machine Learning to Asteroid ClassiFication Utilizing SPectroscoPically Derived SPectroPhotometry DePartment Space Studies Degree Master oF Science In Presenting this thesis in Partial fulfillment of the reQuirements for a graduate degree From the University of North Dakota, I agree that the library of this University shall make it Freely available For insPection. I Further agree that permission For extensive copying For scholarly purposes may be granted by the professor Who suPervised my thesis Work, or in his absence, by the ChairPerson of the department of the dean of the School of Graduate Studies. -

Orbit Determination of Eclipsing Binary Asteroids from Photometry
Orbit determination of eclipsing binary asteroids from photometry Petr Scheirich, Petr Pravec Ondrejov obs., Czech Republic and many observers Orbiting couples: "pas de deux" in the Solar System and the Milky Way Paris, October 10-12, 2011 Lightcurve of ordinary asteroid First binaries resolved from photometry 1994 AW1 (Pravec and Hahn, 1997) First binaries resolved from photometry 1996 FG3 (Pravec et al, 2000) Asynchronous binary Models of binaries derived from photometry • 10 NEA binaries (22 oppositions) • 15 MBA binaries (33 oppositions) Where all the data come from? 2006 Why to do photometry of binaries? Why to do photometry of binaries? • poles distribution • dynamical evolution Lightcurve of binary asteroid Primary Secondary Primary event Secondary event Long-period component extraction L.p. component = mutual events + rotation of secondary (The long period component of) Lightcurve simulation – the direct problem Input parameters: • Heliocentric orbit geometry • Keplerian elements of mutual orbit (circular, eccentric) • Shape and size ratio of components • Scattering law Two-axis ellipsoids or any arbitrary shape approximated by polyhedra with triangular facets The lightcurve of the system is computed using simple ray-traycing code. The inverse problem Fitted parameters: • Keplerian elements of mutual orbit: • a/Ap – semimajor axis • lP – ecl. longitude of orbit’s pole • bP – ecl. latitude of orbit’s pole • Porb – sidereal orbital period • L0 – mean length of secondary at given epoch • e – eccentricity • w – argument of pericenter • Shape and size ratio of components: • flattening of primary Ap /Cp, • elongation of secondary As /Cs, • size ratio of both bodies As / Ap Pre-estimates of initial parameters • Synodic orbital period • Components size ratio Pre-estimates of initial parameters Sidereal orbital period and L0 (argument of mean length of secondary for JD0): Visual identification of contacts: Time-increasing L of contacts should lie on a straight line defined by where n = 2/Psid. -
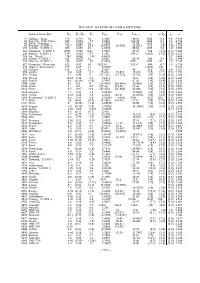
Binary Asteroid Parameters
BINARY ASTEROID PARAMETERS ′ Asteroid/satellite Dp Ds/Dp Ds Perp Pers Perorb a a/Dp ρp a 22 Kalliope/ Linus 170 0.213 36 4.1482 86.16 1065 6.3 2.5 2.910 45 Eugenia/ Petit–Prince 195 0.036 7.0 5.6991 114.38 1184 6.1 1.1 2.724 87 Sylvia/ Romulus 256 0.063 16 5.1836 87.59 1356 5.3 1.5 3.493 90 Antiope/ S/2000 1 86.7 0.955 82.8 16.5051 16.5051 16.5051 171 1.97 1.26 3.154 107 Camilla/ S/2001 1 206 0.050 10 4.8439 89.04 1235 6.0 1.9 3.495 121 Hermione/ S/2002 1 (205) 0.066 (14) 5.5513 61.97 768 (3.7) (1.1) 3.448 130 Elektra/ S/2003 1 179 0.026 4.7 5.225 (94.1) (1252) (7.0) (3.0) 3.124 243 Ida/ Dactyl 28.1 0.048 1.34 4.6336 2.7 2.860 283 Emma/ S/2003 1 145 0.079 11 6.888 80.74 596 4.1 0.8 3.046 379 Huenna/ S/2003 1 90 0.078 7.0 (7.022) 1939 3400 38 1.2 3.136 617 Patroclus/ Menoetius 101 0.92 93 (102.8) 102.8 680 6.7 1.3 5.218 624 Hektor/ Skamandrios 220 0.05 11 6.92051 (1700) (8) 5.242 762 Pulcova 133 0.16 21 5.839 96 810 6.1 1.9 3.157 809 Lundia 6.9 0.89 6.1 15.418 15.418 15.418 (15) (2.2) (2.0) 2.283 854 Frostia 5.7 0.98 6 (37.711) (37.711) 37.711 (24) (4.1) (2.0) 2.368 939 Isberga 10.56 0.29 3.1 2.9173 26.8 (28) (2.6) (2.0) 2.246 1052 Belgica 9.8 (0.36) (3.5) 2.7097 47.26 (38) (3.9) (2.0) 2.236 1089 Tama 9.1 0.9 8 (16.4461) (16.4461) 16.4461 (21) (2.3) (2.0) 2.214 1139 Atami 5 0.8 4.0 (27.45) (27.45) 27.45 (15) (3.1) (2.0) 1.947 1313 Berna 9.5 0.97 9.2 (25.464) (25.464) 25.464 (30) (3.1) (2.0) 2.656 1338 Duponta 7.7 0.24 1.8 3.85453 17.5680 (15) (2.0) (2.0) 2.264 1453 Fennia 6.33 0.28 1.8 4.4121 (23.1) 23.00351 (16) (2.6) (2.0) 1.897 1509 -

Binary Asteroid Population. 2. Anisotropic Distribution of Orbit Poles of Small, Inner Main-Belt Binaries
Binary Asteroid Population. 2. Anisotropic distribution of orbit poles of small, inner main-belt binaries P. Pravec a, P. Scheirich a, D. Vokrouhlick´y b, A. W. Harris c, P. Kuˇsnir´ak a, K. Hornoch a, D. P. Pray d, D. Higgins e, A. Gal´ad a,f, J. Vil´agi f, S.ˇ Gajdoˇs f, L. Kornoˇs f, J. Oey g, M. Hus´arik h, W. R. Cooney i, J. Gross i, D. Terrell i,j, R. Durkee k, J. Pollock ℓ, D. Reichart m, K. Ivarsen m, J. Haislip m, A. LaCluyze m, Yu. N. Krugly n, N. Gaftonyuk o, R. D. Stephens p, R. Dyvig q, V. Reddy r, V. Chiorny n, O. Vaduvescu s,t, P. Longa-Pe˜na s,u, A. Tudorica v,w, B. D. Warner x, G. Masi y, J. Brinsfield z, R. Gon¸calves aa, P. Brown ab, Z. Krzeminski ab, O. Gerashchenko ac, V. Shevchenko n, I. Molotov ad, F. Marchis ae,af aAstronomical Institute, Academy of Sciences of the Czech Republic, Friˇcova 1, CZ-25165 Ondˇrejov, Czech Republic bInstitute of Astronomy, Charles University, Prague, V Holeˇsoviˇck´ach 2, CZ-18000 Prague 8, Czech Republic c4603 Orange Knoll Avenue, La Ca˜nada, CA 91011, U.S.A. dCarbuncle Hill Observatory, W. Brookfield, MA, U.S.A. eHunters Hill Observatory, Ngunnawal, Canberra, Australia f Modra Observatory, Department of Astronomy, Physics of the Earth, and Meteorology, FMFI UK, Bratislava SK-84248, Slovakia gLeura Observatory, Leura, N.S.W., Australia hAstronomical Institute of the Slovak Academy of Sciences, SK-05960 Tatransk´a Lomnica, Slovakia iSonoita Research Observatory, 77 Paint Trail, Sonoita, AZ 85637, U.S.A. -

Catalogue of Minor Planet Names and Discovery Circumstances
Catalogue of Minor Planet Names and Discovery Circumstances Addendum 2006–2008 L.D. Schmadel, Dictionary of Minor Planet Names, DOI 10.1007/978-3-642-01965-4_2, © Springer-Verlag Berlin Heidelberg 2009 Title page of Giuseppe Piazzi’s book “On the discovery of the new planet CERES FERDINANDEA, the eighth of those known in our solar system”. The vignette, against the background of Monte Pelegrini and the city of Palermo, shows an angel observing the goddess Ceres sitting in a carriage drawn by two snakes. The inscription on the telescope “CERES ADDITA COELI” (Ceres was added to the heavens) celebrates this epoch-making discovery of the first minor planets (Courtesy of A. Baldi, Bologne) (29) Amphitrite 15 (29) Amphitrite [2.55, 0.07, 6.1] que l’une de vos lectrices ´etrang`eres, dont la philosophie Discovered 1854 Mar. 1 by A. Marth at London. astronomique enseign´ee par vos ouvrages ´etait devenue (* AN 38, 125) Independently discovered 1854 Mar. la religion, Mlle Antoinette Horneman, est d´ec´ed´ee 2 by N. R. Pogson at Oxford and March 3 by dans le calme d’une conscience ´eclair´ee et tranquille J. Chacornac at Paris. sur l’evolution future, le 14 decembre dernier. Par Named after an Oceanid, wife of Poseidon {see testament authentique et pour aider `a votre œvre planet (4341)} and mother of Triton. (H 5) si utile au progr`es, si d´evou´ee, si d´esinteress´ee, et Named by G. Bishop at whose private South quelle admirait au del`ade toute expression, elle vous Villa Observatory in Regent’s Park the planet was a l´egu’e une somme de cinq mille florins (environ dix discovered. -

Do Slivan States Exist in the Flora Family? I
A&A 546, A72 (2012) Astronomy DOI: 10.1051/0004-6361/201219199 & c ESO 2012 Astrophysics Do Slivan states exist in the Flora family? I. Photometric survey of the Flora region A. Kryszczynska´ 1,F.Colas2,M.Polinska´ 1,R.Hirsch1, V. Ivanova3, G. Apostolovska4, B. Bilkina3, F. P. Velichko5, T. Kwiatkowski1,P.Kankiewicz6,F.Vachier2, V. Umlenski3, T. Michałowski1, A. Marciniak1,A.Maury7, K. Kaminski´ 1, M. Fagas1, W. Dimitrov1, W. Borczyk1, K. Sobkowiak1, J. Lecacheux8,R.Behrend9, A. Klotz10,11, L. Bernasconi12,R.Crippa13, F. Manzini13, R. Poncy14, P. Antonini15, D. Oszkiewicz16,17, and T. Santana-Ros1 1 Astronomical Observatory Institute, Faculty of Physics, Adam Mickiewicz University, Słoneczna 36, 60-286 Poznan,´ Poland e-mail: [email protected] 2 Institut de Mécanique Céleste et Calcul des Éphémérides, Observatoire de Paris, 77 Av. Denfert Rochereau, 75014 Paris, France 3 Institute of Astronomy, Bulgarian Academy of Sciences Tsarigradsko Chausse 72, 1784 Sofia, Bulgaria 4 Faculty of Natural Sciences, Cyril and Methodius University Skopje, Macedonia 5 Institute of Astronomy, Karazin National University, Kharkov, Ukraine 6 Astrophysics Division, Institute of Physics, Jan Kochanowski University, Swietokrzyska´ 15, 25-406 Kielce, Poland 7 San Pedro de Atacama Observatory, Chile 8 Observatoire de Paris, 5 place Jules Janssen, 92195 Meudon, France 9 Geneva Observatory, 1290 Sauverny, Switzerland 10 Institut de Recherche en Astrophysique et Planétologie (IRAP), Université de Toulouse, 9 avenue du colonel Roche, 31028 Toulouse Cedex 4, France 11 Observatoire -

The Minor Planet Bulletin 42 (2015) 237
THE MINOR PLANET BULLETIN OF THE MINOR PLANETS SECTION OF THE BULLETIN ASSOCIATION OF LUNAR AND PLANETARY OBSERVERS VOLUME 42, NUMBER 4, A.D. 2015 OCTOBER-DECEMBER 235. LIGHTCURVE ANALYSIS OF SIX ASTEROIDS Telescope Observatory CCD Refractor 10cm Zonalunar Atik 383L+ Alfonso Carreño Garcerán Zonalunar Observatory (MPC J08) Cassegrain 35cm Isaac Aznar SBIG STL1001E Puzol, Valencia, SPAIN Cassegrain 25cm Vallbona SBIG ST7-XME [email protected] R-Chretien 20cm TRZ QHY8 D-Kirkham 25cm Elche SBIG ST8-XME Amadeo Aznar Macías Isaac Aznar Observatory (MPC Z95) Table 1: List of instruments used for the observations. Aras de los Olmos, Valencia, SPAIN 425 Cornelia is a main-belt asteroid of 64 km discovered by Auguste Charlois in 1896. A total of 546 data points were obtained Enrique Arce Mansego over 4 nights during 2015 March 4-7. The solar phase angle was Vallbona Observatory (MPC J67) +12.0° and +13° at the start and end of the period. Its magnitude Pobla Vallbona, Valencia, SPAIN was V ~ 13.9. The lightcurve shows a period of 17.505 ± 0.001 h Pedro Brines Rodriguez and amplitude of 0.21 mag. The LCDB shows a period of 17.56 h and amplitude of 0.19 mag calculated by Behrend (2004). TRZ Observatory (MPC Z98) Bétera, Valencia, SPAIN Juan Lozano de Haro Elche Observatory (MPC I57) Elche, Alicante, SPAIN (Received: 2015 May 11 Revised: 2015 June 25) Photometric observations of six asteroids were made from 2015 March to May. We report the results of our lightcurve analysis for 425 Cornelia, 625 Xenia, 664 Judith, 785 Bredichina, 910 Anneliese, and 1831 Nicholson. -

Ray, Modern Astronomy
This is a reproduction of a library book that was digitized by Google as part of an ongoing effort to preserve the information in books and make it universally accessible. https://books.google.com I ;r-:-iii \'5.i tfA'K THE PULL MOOH, FBOM A PHOTO&RAPH. ECLECTIC EDUCATIONAL SERIES. THE ELEMENTS ASTRONOMY: COLLEGES, SCHOOLS, AND PRIVATE STUDENTS. WEITTEN FOB THE MATHEMATICAL COURSE OF JOSEPH RAY, M.D., BY SELIM H. PEABODY, M.A., TEACHER OF NATURAL SCIENCES IN TH*! CH'CAGQ BIQK SuHOuL CINCINNATI. WILSON, HINKLE &, CO. PHIL'A: CLAXTON, REMSE*& HAFFELFINGER. NEW YORK : CLARK & MAYNARD. f--3LIC LIBRARY 4787.25 »«T"«, LFMOX ANV F, ICO t, Entered according to Act of Congress, in the year 1869, by WILSON, HINKLE & CO., In the Clerk's Office of the District Court of the United States, for the Southern District of Ohio. ■ • • • • ♦ 1 • • • • « • ELECTROTYPED AT THE FRANKLIN TYPE FOUNDRY. CINCINNATI > g PREFACE. 92 CO C"i In this work I have endeavored to describe and explain the q Mechanism of the Solar System and of the Stellar Universe. ^5 Though written for pupils in the higher grades of public schools, it may be found useful in institutions of still higher rank, and as a foundation for more extended research by the private student. Although well aware that the rigid principles of mathematics and mechanics form the sole foundation of high astronomical attainment, I have carefully avoided abstruse mathematical demonstrations. Most who study Astronomy desire an accurate tiowledge of facts and principles, but need neither for mental - culture nor for practical use such a mastery of methods' as should fit them to become even amateur astronomers.