Structural Analyses of Avocado Sunblotch Viroid Reveal Differences in the Folding of Plus and Minus RNA Strands
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
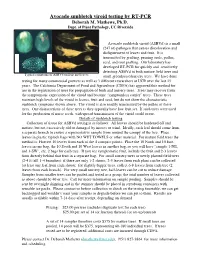
Avocado Sunblotch Viroid Testing by RT-PCR Deborah M
Avocado sunblotch viroid testing by RT-PCR Deborah M. Mathews, Ph.D. Dept. of Plant Pathology, UC Riverside Avocado sunblotch viroid (ASBVd) is a small (247 nt) pathogen that causes discoloration and disfigurement of leaves and fruit. It is transmitted by grafting, pruning tools, pollen, seed, and root grafting. Our laboratory has developed RT-PCR for quickly and sensitively detecting ASBVd in both mature field trees and Typical symptoms of ASBVd on fruit and leaves small greenhouse/nursery trees. We have done testing for many commercial growers as well as 3 different researchers at UCR over the last 15 years. The California Department of Food and Agriculture (CDFA) has approved this method for use in the registration of trees for propagation of buds and nursery trees. Trees may recover from the symptomatic expression of the viroid and become “symptomless carrier” trees. These trees maintain high levels of the viroid in leaves, fruit and seed, but do not show the characteristic sunblotch symptoms shown above. The viroid is also readily transmitted by the pollen of these trees. One characteristic of these trees is they typically have low fruit set. If such trees were used for the production of nurse seeds, widespread transmission of the viroid could occur. Details of sunblotch testing Collection of tissue for ASBVd testing is as follows: All leaves should be hardened off and mature, but not excessively old or damaged by insects or wind. Ideally, each leaf should come from a separate branch to ensure a representative sample from around the canopy of the tree. Place leaves in plastic ziplock bags with NO WET TOWELS or other material. -

Molecular Variability of Apple Hammerhead Viroid from Italian
Virus Research 270 (2019) 197644 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres Short communication Molecular variability of apple hammerhead viroid from Italian apple varieties supports the relevance in vivo of its branched conformation T stabilized by a kissing loop interaction Michela Chiumentia, Beatriz Navarroa, Pasquale Veneritob, Francesco Civitac, ⁎ ⁎ Angelantonio Minafraa, , Francesco Di Serioa, a Istituto per la Protezione Sostenibile delle Piante (CNR), Bari, Italy b Centro di Ricerca, Sperimentazione e Formazione in Agricoltura “Basile Caramia”, Locorotondo, Italy c SINAGRI – Università degli Studi di Bari “Aldo Moro”, Bari, Italy ARTICLE INFO ABSTRACT Keywords: In the absence of protein-coding ability, viroid RNAs rely on direct interactions with host factors for their Hammerhead infectivity. RNA structural elements are likely involved in these interactions. Therefore, preservation of a Non-Coding RNA structural element, despite the sequence variability existing between the variants of a viroid population, is Sequence variability considered a solid evidence of its relevant role in vivo. In this study, apple hammerhead viroid (AHVd) was first Secondary structure identified in the two apple cultivars ‘Mela Rosa Guadagno’ (MRG) and ‘Agostinella’ (AG), which are cultivated Co-Variation since long in Southern Italy, thus providing the first solid evidence of its presence in this country. Then, the Stem-Loop natural variability of AHVd viroid populations infecting MRG and AG was studied. The sequence variants from the two Italian isolates shared only 82.1–87.7% sequence identity with those reported previously from other geographic areas, thus providing the possibility of exploring the impact of this sequence divergence on the proposed secondary structure. -

Detection of Avocado Sunblotch Viroid and Estimation of Infection Among Accessions in the National Germplasm Collection for Avocado
Proc. Fla. State Hort. Soc. 109:235-237. 1996. DETECTION OF AVOCADO SUNBLOTCH VIROID AND ESTIMATION OF INFECTION AMONG ACCESSIONS IN THE NATIONAL GERMPLASM COLLECTION FOR AVOCADO Catherine M. Running and Raymond J. Schnell National Germplasm Repository U. S. Department of Agriculture Agricultural Research Service 13601 Old Cutler Road, Miami, FL 33158 David N. Kuhn Florida International University Dept. of Biological Sciences Miami, FL 33199 Additional index words. Persea Americana, RT-PCR, viroid, DNA, RNA, sequence variation, germplasm. ABSTRACT Reverse transcription-polymerase chain reaction (RTPCR) was used to determine the incidence of infection by avocado sunblotch viroid (ASBVd) in the germplasm collection at the National Germplasm Repository at Miami (NGR-Mia). Of the 429 avocado plants growing at the repository, 81 (18.9%) are infected with the viroid. The 429 plants represent 237 accessions. There are multiple plants of some accessions and for 42 accessions (17%) every plant is infected with the viroid. There was no apparent relationship between host race and the incidence of infection. Symptoms of sunblotch disease on avocado (Persea Americana Mill.) manifest as a general decline of tree vigor with sunken, yellow areas on the fruit that lessen its marketability. The disease was first described as infectious, rather than physiological, by Home and Parker (1931). The infectious agent has since been determined to be an RNA viroid known as Avocado Sunblotch Viroid (ASBVd) (Dale and Allen, 1979; Thomas and Mohammed, 1979). The viroid is transmitted by budding and grafting, including natural root grafting (Home etal, 1941; Whitsell, 1952), seed (Wallace and Drake, 1962), and pollen (Desjardins et al., 1979). -

THE MOLECULAR CHARACTERIZATION of the VIRUS and VIRUS-LIKE AGENTS PRESENT in TA TAO 5 GERMPLASM of PRUNUS PERSICA Diana Marini Clemson University, [email protected]
Clemson University TigerPrints All Dissertations Dissertations 12-2007 THE MOLECULAR CHARACTERIZATION OF THE VIRUS AND VIRUS-LIKE AGENTS PRESENT IN TA TAO 5 GERMPLASM OF PRUNUS PERSICA Diana Marini Clemson University, [email protected] Follow this and additional works at: https://tigerprints.clemson.edu/all_dissertations Part of the Agronomy and Crop Sciences Commons Recommended Citation Marini, Diana, "THE MOLECULAR CHARACTERIZATION OF THE VIRUS AND VIRUS-LIKE AGENTS PRESENT IN TA TAO 5 GERMPLASM OF PRUNUS PERSICA " (2007). All Dissertations. 143. https://tigerprints.clemson.edu/all_dissertations/143 This Dissertation is brought to you for free and open access by the Dissertations at TigerPrints. It has been accepted for inclusion in All Dissertations by an authorized administrator of TigerPrints. For more information, please contact [email protected]. THE MOLECULAR CHARACTERIZATION OF THE VIRUS AND VIRUS-LIKE AGENTS PRESENT IN TA TAO 5 GERMPLASM OF PRUNUS PERSICA A Dissertation Presented to the Graduate School of Clemson University In Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Plant and Environmental Sciences by Diana Beatriz Marini December 2007 Accepted by: Dr. Simon W. Scott, Committee Chair Dr. N. Dwight Camper Dr. Steven N. Jeffers Dr. Gregory L. Reighard ABSTRACT Peach production in the southeastern United States is limited by late spring freezes. Ta Tao 5 germplasm, used either as an interstem or by chip bud inoculation, has been shown to delay bloom and avoid the effects of these late freezes. The growth modification is graft transmissible and the germplasm has been found to be infected with ACLSV, APruV-3, and PLMVd. Using a combination of PCR, cloning, and sequencing techniques, a molecular characterization of the three graft-transmissible agents present in Ta Tao 5 has been completed. -

Replication of Avocado Sunblotch Viroid in the Cyanobacterium Nostoc Sp. PCC 7120
atholog P y & nt a M Latifi et al., J Plant Pathol Microbiol 2016, 7:4 l i P c f r o o DOI: 10.4172/2157-7471.1000341 b l i Journal of a o l n o r g u y o J ISSN: 2157-7471 Plant Pathology & Microbiology Research Article Open Access Replication of Avocado Sunblotch Viroid in the Cyanobacterium Nostoc Sp. PCC 7120 Amel Latifi1*, Christophe Bernard1, Laura da Silva2, Yannick Andéol3, Amine Elleuch4, Véronique Risoul1, Jacques Vergne2 and Marie-Christine Maurel2 1Aix Marseille University, CNRS, UMR Chemistry Laboratory Bacterial 7283. 31 Chemin Joseph Aiguier 13009 Marseille Cedex 20, France 2Institute of Systematics, Evolution, Biodiversity ( ISyEB ), CNRS, MNHN , UPMC, EPHE, UPMC Sorbonne University, 57 rue Cuvier, PO Box 50, F-75005 Paris, France 3Team Enzymology of RNA, UR6, UPMC Sorbonne University, 75252 Paris Cedex 05 France 4Laboratory of Plant Biotechnology, Faculty of Sciences of Sfax, University of Sfax, BP 1171, 3000 Sfax, Tunisia Abstract Viroids are small infectious RNA molecules that replicate in plants via RNA-RNA replication processes. The molecular mechanism responsible for this replication has attracted great interest, and studies on this topic have yielded interesting biological findings on the processes in which RNA is involved. Viroids belonging to the Avsunviroidae family replicate in the chloroplasts of infected hosts. It has by now been established that chloroplasts and cyanobacteria share a common have ancestor. In view of this phylogenetic relationship, we investigated whether a member of the Avsunviroidae family could be replicated in a cyanobacterium. The results obtained here show that Avocado Sunblotch Viroid (ASBVd) RNA is able to replicate in the filamentous cyanobacterium Nostoc PCC 7120. -

Hammerhead Ribozymes Against Virus and Viroid Rnas
Hammerhead Ribozymes Against Virus and Viroid RNAs Alberto Carbonell, Ricardo Flores, and Selma Gago Contents 1 A Historical Overview: Hammerhead Ribozymes in Their Natural Context ................................................................... 412 2 Manipulating Cis-Acting Hammerheads to Act in Trans ................................. 414 3 A Critical Issue: Colocalization of Ribozyme and Substrate . .. .. ... .. .. .. .. .. ... .. .. .. .. 416 4 An Unanticipated Participant: Interactions Between Peripheral Loops of Natural Hammerheads Greatly Increase Their Self-Cleavage Activity ........................... 417 5 A New Generation of Trans-Acting Hammerheads Operating In Vitro and In Vivo at Physiological Concentrations of Magnesium . ...... 419 6 Trans-Cleavage In Vitro of Short RNA Substrates by Discontinuous and Extended Hammerheads ........................................... 420 7 Trans-Cleavage In Vitro of a Highly Structured RNA by Discontinuous and Extended Hammerheads ........................................... 421 8 Trans-Cleavage In Vivo of a Viroid RNA by an Extended PLMVd-Derived Hammerhead ........................................... 422 9 Concluding Remarks and Outlooks ........................................................ 424 References ....................................................................................... 425 Abstract The hammerhead ribozyme, a small catalytic motif that promotes self- cleavage of the RNAs in which it is found naturally embedded, can be manipulated to recognize and cleave specifically -

1 Method for the Detection of Pospiviroids on Tomato Seed Crop
Method for the Detection of Pospiviroids on Tomato Seed Crop Tomato (Solanum lycopersicum) Pathogens Pospiviroidae: Citrus exocortis viroid (CEVd), Columnea latent viroid (CLVd), Mexican papita viroid (MPVd), Pepper chat fruit viroid (PCFVd), Potato spindle tuber viroid (PSTVd), Tomato apical stunt viroid (TASVd), Tomato chlorotic dwarf viroid (TCDVd), Tomato planta macho viroid (TPMVd) ISHI-Veg recommends using the Naktuinbouw protocol Sample and sub-sample sizes The Naktuinbouw protocol uses 3,000 or 20,000 seeds in the sample. A sample size of 3,000 seeds with a maximum sub-sample size of 1,000 seeds is adequate for detecting pospiviroidae in tomato seed. NOTE: An important factor that determines the sample size of the seed to be tested is the epidemiology of the disease, i.e. 1. the rate of disease transmission from infected seed to seedling and 2. the spread of the disease in a seed production unit 1. Rate of disease transmission Several studies have experimentally demonstrated variable rates of transmission of pospiviroids from infected seed to seedlings (Kryczynski et al., 1988 for PSTVd, Singh and Dilworth, 2009 for TCDVd, Antignus et al., 2007 for TASVd). However, there are also studies in which no seed transmission was observed (Singh et al., 1999 and Matsushita et al., 2011 for TCDVd). In all these studies the infected seeds were obtained by artificially infecting mother plants. In the few studies using naturally infected seed lots, transmission rates are very low; 0,08 % for TCDVd based on 2500 seeds (Candresse et al., 2010) and 0,27% for PSTVd based on 370 seeds (Brunschot et al., 2014). -

Recognizing Avocado Sunblotch Disease
RECOGNIZING AVOCADO SUNBLOTCH DISEASE 1J. A. Dodds, 1D Mathews, 2M. L. Arpaia and 3G. W. Witney. 1Dept. Plant Pathology, University of California, Riverside, CA 92521. A. 2Dept. Botany and Plant Sciences, University of California, Riverside, CA 92521. 3California Avocado Commission, 1251 E. Dyer Rd., Suite 210, Santa Ana, CA 92705. Avocado sunblotch is a disease which has been known for over 60 years. Sunblotch is characterized by abnormal tree growth, reduced yield and a high proportion of small or misshapen fruit. Specific symptoms may be observed on fruit, leaves and stems. Fruit may show overall distortion, sometimes with sunken yellow and red depressions on the surface. These fruit are graded as standards and receive lower returns from the packinghouse (Figure A). Foliar symptoms can include a general thinning of B. the canopy, with individual leaves showing bleaching (white patches), variegation (yellow and green patches), and distortion (Figure B, C). Stems may also show discoloration with yellow, white or pink streaks (Figure D), and in extreme cases may be completely chlorotic (yellow/white). Infected trees often appear stunted and somewhat sprawling. It is possible for trees to “recover” from the disease. The “recovered” tree will have no apparent visual disease symptoms but it still carries the viroid. These trees are termed “symptomless” carriers. Such trees typically have very low fruit yield or at times may set heavy crops of small fruit. If symptomless trees are topworked with disease-free material, the topworked material will become infected and can exhibit classical sunblotch symptoms revealing C. the presence of sunblotch. If a symptomless tree is subject to a stress such as fire or is stumped, the regrowth may once again exhibit sunblotch symptoms. -

Homology-Independent Discovery of Replicating Pathogenic Circular Rnas by Deep Sequencing and a New Computational Algorithm
Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm Qingfa Wua,b,1, Ying Wangb,1, Mengji Caob, Vitantonio Pantaleoc, Joszef Burgyanc, Wan-Xiang Lib, and Shou-Wei Dingb,2 aSchool of Life Sciences, University of Science and Technology of China, Hefei, 230027, China; bDepartment of Plant Pathology and Microbiology and Institute for Integrative Genome Biology, University of California, Riverside, CA 92521; and cIstituto di Virologia Vegetale, Consiglio Nazionale delle Ricerche, Torino, Italy Edited by George E. Bruening, University of California, Davis, CA, and approved January 24, 2012 (received for review October 28, 2011) A common challenge in pathogen discovery by deep sequencing Application of deep sequencing technologies has facilitated approaches is to recognize viral or subviral pathogens in samples discovery of viruses by purification- and culture-independent of diseased tissue that share no significant homology with a known approaches. In metagenomic approaches, viral sequences are pathogen. Here we report a homology-independent approach for enriched by partial purification of viral particles before deep se- discovering viroids, a distinct class of free circular RNA subviral quencing (6, 7). In contrast, enrichment of viral sequences is pathogens that encode no protein and are known to infect plants achieved by isolating the fraction of small RNAs 20–30 nt in only. Our approach involves analyzing the sequences of the total length for deep sequencing in virus discovery by deep sequencing small RNAs of the infected plants obtained by deep sequencing and assembly of total host small RNAs (vdSAR) (8, 9). Pro- with a unique computational algorithm, progressive filtering of duction of virus-derived small interfering RNAs (siRNAs) is a key overlapping small RNAs (PFOR). -

The Avocado Sunblotch Viroid: an Invisible Foe of Avocado
viruses Review The Avocado Sunblotch Viroid: An Invisible Foe of Avocado José Ramón Saucedo Carabez 1,*, Daniel Téliz Ortiz 2 , Moisés Roberto Vallejo Pérez 3 and Hugo Beltrán Peña 4 1 Departamento de Investigación Aplicada-Driscolls, Libramiento Sur #1620, Jacona 59833, Michoacán, Mexico 2 Postgrado en Fitosanidad-Fitopatología, Colegio de Postgraduados, Km. 36.5, Montecillo, Texcoco 56230, Estado de México, Mexico; [email protected] 3 Consejo Nacional de Ciencia y Tecnología-Universidad Autónoma de San Luis Potosí, Álvaro Obregón #64, San Luis Potosí 78000, San Luis Potosí, Mexico; [email protected] 4 Departamento de Ciencias Naturales y Exactas-Fitopatología, Universidad Autónoma de Occidente UR Los Mochis, Boulevard Macario Gaxiola, Los Mochis 81223, Sinaloa, Mexico; [email protected] * Correspondence: [email protected] Received: 1 May 2019; Accepted: 25 May 2019; Published: 29 May 2019 Abstract: This review collects information about the history of avocado and the economically important disease, avocado sunblotch, caused by the avocado sunblotch viroid (ASBVd). Sunblotch symptoms are variable, but the most common in fruits are irregular sunken areas of white, yellow, or reddish color. On severely affected fruits, the sunken areas may become necrotic. ASBVd (type species Avocado sunblotch viroid, family Avsunviroidae) replicates and accumulates in the chloroplast, and it is the smallest plant pathogen. This pathogen is a circular single-stranded RNA of 246–251 nucleotides. ASBVd has a restricted host range and only few plant species of the family Lauraceae have been confirmed experimentally as additional hosts. The most reliable method to detect ASBVd in the field is to identify symptomatic fruits, complemented in the laboratory with reliable and sensitive molecular techniques to identify infected but asymptomatic trees. -

Viroid-2018 Book of Abstracts
Viroid-2018 Viroid-2018 International Conference on Viroids and Viroid-Like RNAs 5-7 July 2018, Valencia, Spain Polytechnic City of Innovation, Universitat Politècnica de València Avenida de los Naranjos, 46022 Valencia, Spain http://www.ibmcp.upv.es/viroid-2018/ [email protected] Book of Abstracts Organizer: José-Antonio Daròs Instituto de Biología Molecular y Celular de Plantas (IBMCP) CSIC-Universitat Politècnica de Valencia Avenida de los Naranjos, 46022Valencia, Spain [email protected] http://personales.upv.es/jadaros/ Updated version, 3 July 2018 1 Viroid-2018 INDEX Supporting institutions and contributing companies ……………… 3 Conference venue …………………………………….……….……… 4 Conference program ………………………………………………… 6 List of participants ………………………………………...………… 12 Abstracts, oral presentations …………….………….……….……… 16 Abstracts, poster presentations …………….………….……….…… 47 Index of authors ……………………………………………………… 70 Notes …………………………..……………………….……………… 72 2 Viroid-2018 SUPPORTING INSTITUTIONS Grants from Ministerio de Ciencia, Innovación y Universidades (Spain): BIO2014-54269-R, BIO2017-83184-R and BIO2017-91865-EXP Grant from Generalitat Valenciana: AORG/2018/114 CONTRIBUTING COMPANIES Biotest Diagnosticos S.L., http://www.biotestdiagnosticos.es/ Cultek, http://www.cultek.com/ Durviz, http://durviz.com/ Épica, http://epicasl.com/ Fisher Scientific, http://www.fishersci.com GenoChem, http://geno-chem.com/ IDT, http://www.idtdna.com/pages NZYTech, http://www.nzytech.com/ Werfen, http://es.werfen.com/ VWR, http://www.vwr.com/ 3 Viroid-2018 CONFERENCE VENUE Campus Universitat Politècnica de Valencia, corner between Avenida de los Naranjos and Calle Ingeniero Fausto Elio, Access J, Ciudad Politécnica de la Innovación (CPI), Red Prism. 4 Viroid-2018 5 Viroid-2018 CONFERENCE PROGRAM Thursday, July 5th 09:00-10:00. Registration at CPI Red Prism 10:00-10:20. Opening Chairman: Robert A. -

Pospiviroidae Viroids in Naturally Infected Stone and Pome Fruits In
21st International Conference on Virus and other Graft Transmissible Diseases of Fruit Crops Pospiviroidae viroids in naturally infected stone and pome fruits in Greece Kaponi, M.S.1, Luigi, M.2, Barba, M.2, Kyriakopoulou, P.E.I I Agricultural University of Athens, Iera Odos 75, 11855 Athens, Greece 2 CRA-PAV, Centro di Ricerca per la Patologia Vegeta le, 00156 Rome, Italy Abstract Viroid research on pome and stone fruit trees in Greece is important, as it seems that such viroids are widespread in the country and may cause serious diseases. Our research dealt with three Pospiviroidae species infecting pome and stone fruit trees, namely Apple scar skin viroid (ASSVd), Pear blister canker viroid (PBCVd) and Hop stunt viroid (HSVd). Tissue-print hybridization, reverse transcription-polymerase chain reaction (RT-PCR), cloning and sequencing techniques were successfully used for the detection and identification of these viroids in a large number of pome and stone fruit tree samples from various areas of Greece (Peloponnesus, Macedonia, Thessaly, Attica and Crete). The 58 complete viroid sequences obtained (30 ASSVd, 16 PBCVd and 12 HSVd) were submitted to the Gen Bank. Our results showed the presence of ASSVd in apple, pear, wild apple (Malus sylvestris), wild pear (Pyrus amygdaliformis) and sweet cherry; HSVd in apricot, peach, plum, sweet cherry, bullace plum (Prunus insititia), apple and wild apple; and PBCVd in pear, wild pear, quince, apple and wild apple. This research confirmed previous findings of infection of Hellenic apple, pear and wild pear with ASSVd, pear, wild pear and quince with PBCVd and apricot with HSVd.