Non-Monophyly and Intricate Morphological Evolution Within The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
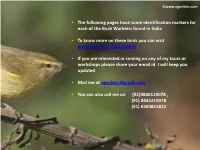
• the Following Pages Have Some Identification Markers for Each of the Bush Warblers Found in India
©www.ogaclicks.com • The following pages have some identification markers for each of the Bush Warblers found in India • To know more on these birds you can visit www.ogaclicks.com/warbler • If you are interested in coming on any of my tours or workshops please share your email id. I will keep you updated • Mail me at [email protected] • You can also call me on (91)9840119078 , (91) 9445219078 (91) 6369815812 Abberant Bush Warbler Identification Tips - Nominate Abberant Bush Warbler : Cettia flavolivacea : Resident of Himalayas from North Central India (East of Himachal Pradesh and Uttarakhand) Crown is plain brown Pale yellowish supercilium Bill is dark horn- Dark eyestripe brown, pale pink Upperparts are yellowish base of lower Brown Ear-coverts olive-green mandible Narrow whitish eyering Throat is unspotted whitish Breast is darker olive Dull olive-yellow undertail-coverts Buffish or olive- yellow Underparts Flanks are darker olive Legs are yellow to dusky pinkish-brown ©www.ogaclicks.com Reference : www.HBW.com Brown Bush Warbler Identification Tips - Nominate Brown Bush Warbler : Bradypterus luteoventris : Resident of North East India (from Darjeeling, in West Bengal, Eastwards to Arunachal Pradesh and Nagaland) Crown is plain brown Deep buff supercilium upper mandible is Brown eyestripe blackish-brown, lower mandible Brown Ear-coverts fleshy-yellow with blackish-brown tip Upperparts are plain brown Throat is unspotted whitish Breast is Brown Belly is unspotted whitish Deep buff undertail-coverts Deep buff Flanks Legs are flesh-brown -

Bird Diversity in Northern Myanmar and Conservation Implications
ZOOLOGICAL RESEARCH Bird diversity in northern Myanmar and conservation implications Ming-Xia Zhang1,2, Myint Kyaw3, Guo-Gang Li1,2, Jiang-Bo Zhao4, Xiang-Le Zeng5, Kyaw Swa3, Rui-Chang Quan1,2,* 1 Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin Nay Pyi Taw 05282, Myanmar 2 Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla Yunnan 666303, China 3 Hponkan Razi Wildlife Sanctuary Offices, Putao Kachin 01051, Myanmar 4 Science Communication and Training Department, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla Yunnan 666303, China 5 Yingjiang Bird Watching Society, Yingjiang Yunnan 679300, China ABSTRACT Since the 1990s, several bird surveys had been carried out in the Putao area (Rappole et al, 2011). Under the leadership of We conducted four bird biodiversity surveys in the the Nature and Wildlife Conservation Division (NWCD) of the Putao area of northern Myanmar from 2015 to 2017. Myanmar Forestry Ministry, two expeditions were launched in Combined with anecdotal information collected 1997–1998 (Aung & Oo, 1999) and 2001–2009 (Rappole et al., between 2012 and 2015, we recorded 319 bird 2011), providing the most detailed inventory of local avian species, including two species (Arborophila mandellii diversity thus far. 1 and Lanius sphenocercus) previously unrecorded in Between December 2015 and May 2017, the Southeast Asia Myanmar. Bulbuls (Pycnonotidae), babblers (Timaliidae), Biodiversity Research Institute, Chinese Academy of Sciences pigeons and doves (Columbidae), and pheasants (CAS-SEABRI), Forest Research Institute (FRI) of Myanmar, and partridges (Phasianidae) were the most Hponkan Razi Wildlife Sanctuary (HPWS), and Hkakabo Razi abundant groups of birds recorded. -

Disaggregation of Bird Families Listed on Cms Appendix Ii
Convention on the Conservation of Migratory Species of Wild Animals 2nd Meeting of the Sessional Committee of the CMS Scientific Council (ScC-SC2) Bonn, Germany, 10 – 14 July 2017 UNEP/CMS/ScC-SC2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II (Prepared by the Appointed Councillors for Birds) Summary: The first meeting of the Sessional Committee of the Scientific Council identified the adoption of a new standard reference for avian taxonomy as an opportunity to disaggregate the higher-level taxa listed on Appendix II and to identify those that are considered to be migratory species and that have an unfavourable conservation status. The current paper presents an initial analysis of the higher-level disaggregation using the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World Volumes 1 and 2 taxonomy, and identifies the challenges in completing the analysis to identify all of the migratory species and the corresponding Range States. The document has been prepared by the COP Appointed Scientific Councilors for Birds. This is a supplementary paper to COP document UNEP/CMS/COP12/Doc.25.3 on Taxonomy and Nomenclature UNEP/CMS/ScC-Sc2/Inf.3 DISAGGREGATION OF BIRD FAMILIES LISTED ON CMS APPENDIX II 1. Through Resolution 11.19, the Conference of Parties adopted as the standard reference for bird taxonomy and nomenclature for Non-Passerine species the Handbook of the Birds of the World/BirdLife International Illustrated Checklist of the Birds of the World, Volume 1: Non-Passerines, by Josep del Hoyo and Nigel J. Collar (2014); 2. -

Passeriformes: Cisticolidae: Orthotomus) from the Mekong Floodplain of Cambodia
FORKTAIL 29 (2013): 1–14 http://zoobank.org/urn:lsid:zoobank.org:pub:F1778491-B6EE-4225-95B2-2843B32CBA08 A new species of lowland tailorbird (Passeriformes: Cisticolidae: Orthotomus) from the Mekong floodplain of Cambodia S. P. MAHOOD, A. J. I. JOHN, J. C. EAMES, C. H. OLIVEROS, R. G. MOYLE, HONG CHAMNAN, C. M. POOLE, H. NIELSEN & F. H. SHELDON Based on distinctive morphological and vocal characters we describe a new species of lowland tailorbird Orthotomus from dense humid lowland scrub in the floodplain of the Mekong, Tonle Sap and Bassac rivers of Cambodia. Genetic data place it in the O. atrogularis–O. ruficeps–O. sepium clade. All data suggest that the new species is most closely related to O. atrogularis, from which genetic differences are apparently of a level usually associated with subspecies. However the two taxa behave as biological species, existing locally in sympatry and even exceptionally in syntopy, without apparent hybridisation. The species is known so far from a small area within which its habitat is declining in area and quality. However, although birds are found in a number of small habitat fragments (including within the city limits of Phnom Penh), most individuals probably occupy one large contiguous area of habitat in the Tonle Sap floodplain. We therefore recommend it is classified as Near Threatened on the IUCN Red List. The new species is abundant in suitable habitat within its small range. Further work is required to understand more clearly the distribution and ecology of this species and in particular its evolutionary relationship with O. atrogularis. INTRODUCTION and its major tributaries (Duckworth et al. -

Bird List Column A: We Should Encounter (At Least a 90% Chance) Column B: May Encounter (About a 50%-90% Chance) Column C: Possible, but Unlikely (20% – 50% Chance)
THE PHILIPPINES Prospective Bird List Column A: we should encounter (at least a 90% chance) Column B: may encounter (about a 50%-90% chance) Column C: possible, but unlikely (20% – 50% chance) A B C Philippine Megapode (Tabon Scrubfowl) X Megapodius cumingii King Quail X Coturnix chinensis Red Junglefowl X Gallus gallus Palawan Peacock-Pheasant X Polyplectron emphanum Wandering Whistling Duck X Dendrocygna arcuata Eastern Spot-billed Duck X Anas zonorhyncha Philippine Duck X Anas luzonica Garganey X Anas querquedula Little Egret X Egretta garzetta Chinese Egret X Egretta eulophotes Eastern Reef Egret X Egretta sacra Grey Heron X Ardea cinerea Great-billed Heron X Ardea sumatrana Purple Heron X Ardea purpurea Great Egret X Ardea alba Intermediate Egret X Ardea intermedia Cattle Egret X Ardea ibis Javan Pond-Heron X Ardeola speciosa Striated Heron X Butorides striatus Yellow Bittern X Ixobrychus sinensis Von Schrenck's Bittern X Ixobrychus eurhythmus Cinnamon Bittern X Ixobrychus cinnamomeus Black Bittern X Ixobrychus flavicollis Black-crowned Night-Heron X Nycticorax nycticorax Western Osprey X Pandion haliaetus Oriental Honey-Buzzard X Pernis ptilorhynchus Barred Honey-Buzzard X Pernis celebensis Black-winged Kite X Elanus caeruleus Brahminy Kite X Haliastur indus White-bellied Sea-Eagle X Haliaeetus leucogaster Grey-headed Fish-Eagle X Ichthyophaga ichthyaetus ________________________________________________________________________________________________________ WINGS ● 1643 N. Alvernon Way Ste. 109 ● Tucson ● AZ ● 85712 ● www.wingsbirds.com -

Indonesia Highlights of Western Indonesia (Flores, Komodo, Bali, Java & Sumatra) 15Th to 28Th July 2019 (14 Days)
Indonesia Highlights of Western Indonesia (Flores, Komodo, Bali, Java & Sumatra) 15th to 28th July 2019 (14 days) Trip Report Javan Banded Pitta by Glen Valentine Trip report compiled by Tour Leader: Glen Valentine Top 10 list as voted for by the tour participants: 1. Javan Trogon 2. Red-crowned Barbet 3. Green Broadbill 4. Javan Frogmouth 5. Buffy Fish Owl 6. Pygmy Cupwing 7. Rufous-collared Kingfisher 8. Javan Banded Pitta 9. Red-bearded Bee-eater 10. Bali Myna Bali Myna (Starling) by Dennis Braddy Tour Summary… This short but extremely productive and varied tour, covering a fine selection of hand-picked “top birding sites and destinations” throughout Western Indonesia was an immense success, once again and was an absolute joy to lead due to our enthusiastic, fun and very good-natured group. Our quick-fire, two-week tour of western Indonesia, kicked off in Denpasar, on the island of Bali where we all met up at the Harris Hotel for an introductory dinner and flight the following morning to the island of Flores, situated in Nusa Tenggara (The Lesser Sundas), a chain of islands running mostly east/west to the east of Wallace’s line, therefore having a distinctly Australasian flair about their avifauna. After arriving in the large, coastal town of Labuan Bajo, the gateway to the popular and famous Komodo Island, we boarded our minibus and began the windy drive east, up into the hills, towards our first biding locality of the tour, the forest reserve of Puarlolo. This small reserve was initially set aside to protect the endemic and highly threatened Flores Monarch that was only discovered from this area as recently as 1971 and is still only known from a few scattered localities in the sub-montane forest on Flores. -

Moluccas 15 July to 14 August 2013 Henk Hendriks
Moluccas 15 July to 14 August 2013 Henk Hendriks INTRODUCTION It was my 7th trip to Indonesia. This time I decided to bird the remote eastern half of this country from 15 July to 14 August 2013. Actually it is not really a trip to the Moluccas only as Tanimbar is part of the Lesser Sunda subregion, while Ambon, Buru, Seram, Kai and Boano are part of the southern group of the Moluccan subregion. The itinerary I made would give us ample time to find most of the endemics/specialties of the islands of Ambon, Buru, Seram, Tanimbar, Kai islands and as an extension Boano. The first 3 weeks I was accompanied by my brother Frans, Jan Hein van Steenis and Wiel Poelmans. During these 3 weeks we birded Ambon, Buru, Seram and Tanimbar. We decided to use the services of Ceisar to organise these 3 weeks for us. Ceisar is living on Ambon and is the ground agent of several bird tour companies. After some negotiations we settled on the price and for this Ceisar and his staff organised the whole trip. This included all transportation (Car, ferry and flights), accommodation, food and assistance during the trip. On Seram and Ambon we were also accompanied by Vinno. You have to understand that both Ceisar and Vinno are not really bird guides. They know the sites and from there on you have to find the species yourselves. After these 3 weeks, Wiel Poelmans and I continued for another 9 days, independently, to the Kai islands, Ambon again and we made the trip to Boano. -

Bird Checklists of the World Country Or Region: Myanmar
Avibase Page 1of 30 Col Location Date Start time Duration Distance Avibase - Bird Checklists of the World 1 Country or region: Myanmar 2 Number of species: 1088 3 Number of endemics: 5 4 Number of breeding endemics: 0 5 Number of introduced species: 1 6 7 8 9 10 Recommended citation: Lepage, D. 2021. Checklist of the birds of Myanmar. Avibase, the world bird database. Retrieved from .https://avibase.bsc-eoc.org/checklist.jsp?lang=EN®ion=mm [23/09/2021]. Make your observations count! Submit your data to ebird. -

An Update of Wallacels Zoogeographic Regions of the World
REPORTS To examine the temporal profile of ChC produc- specification of a distinct, and probably the last, 3. G. A. Ascoli et al., Nat. Rev. Neurosci. 9, 557 (2008). tion and their correlation to laminar deployment, cohort in this lineage—the ChCs. 4. J. Szentágothai, M. A. Arbib, Neurosci. Res. Program Bull. 12, 305 (1974). we injected a single pulse of BrdU into pregnant A recent study demonstrated that progeni- CreER 5. P. Somogyi, Brain Res. 136, 345 (1977). Nkx2.1 ;Ai9 females at successive days be- tors below the ventral wall of the lateral ventricle 6. L. Sussel, O. Marin, S. Kimura, J. L. Rubenstein, tween E15 and P1 to label mitotic progenitors, (i.e., VGZ) of human infants give rise to a medial Development 126, 3359 (1999). each paired with a pulse of tamoxifen at E17 to migratory stream destined to the ventral mPFC 7. S. J. Butt et al., Neuron 59, 722 (2008). + 18 8. H. Taniguchi et al., Neuron 71, 995 (2011). label NKX2.1 cells (Fig. 3A). We first quanti- ( ). Despite species differences in the develop- 9. L. Madisen et al., Nat. Neurosci. 13, 133 (2010). fied the fraction of L2 ChCs (identified by mor- mental timing of corticogenesis, this study and 10. J. Szabadics et al., Science 311, 233 (2006). + phology) in mPFC that were also BrdU+. Although our findings raise the possibility that the NKX2.1 11. A. Woodruff, Q. Xu, S. A. Anderson, R. Yuste, Front. there was ChC production by E15, consistent progenitors in VGZ and their extended neurogenesis Neural Circuits 3, 15 (2009). -

GHANA MEGA Rockfowl & Upper Guinea Specials Th St 29 November to 21 December 2011 (23 Days)
GHANA MEGA Rockfowl & Upper Guinea Specials th st 29 November to 21 December 2011 (23 days) White-necked Rockfowl by Adam Riley Trip Report compiled by Tour Leader David Hoddinott RBT Ghana Mega Trip Report December 2011 2 Trip Summary Our record breaking trip total of 505 species in 23 days reflects the immense birding potential of this fabulous African nation. Whilst the focus of the tour was certainly the rich assemblage of Upper Guinea specialties, we did not neglect the interesting diversity of mammals. Participants were treated to an astonishing 9 Upper Guinea endemics and an array of near-endemics and rare, elusive, localized and stunning species. These included the secretive and rarely seen White-breasted Guineafowl, Ahanta Francolin, Hartlaub’s Duck, Black Stork, mantling Black Heron, Dwarf Bittern, Bat Hawk, Beaudouin’s Snake Eagle, Congo Serpent Eagle, the scarce Long-tailed Hawk, splendid Fox Kestrel, African Finfoot, Nkulengu Rail, African Crake, Forbes’s Plover, a vagrant American Golden Plover, the mesmerising Egyptian Plover, vagrant Buff-breasted Sandpiper, Four-banded Sandgrouse, Black-collared Lovebird, Great Blue Turaco, Black-throated Coucal, accipiter like Thick- billed and splendid Yellow-throated Cuckoos, Olive and Dusky Long-tailed Cuckoos (amongst 16 cuckoo species!), Fraser’s and Akun Eagle-Owls, Rufous Fishing Owl, Red-chested Owlet, Black- shouldered, Plain and Standard-winged Nightjars, Black Spinetail, Bates’s Swift, Narina Trogon, Blue-bellied Roller, Chocolate-backed and White-bellied Kingfishers, Blue-moustached, -

The Avifauna of the Barito Ulu Region, Central Kalimantan
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by KUKILA KUKILA 5 No. 2 (1991): 99 -116 THE AVIFAUNA OF THE BARITO ULU REGION, CENTRAL KALIMANTAN by Roger Wilkinson, Guy Dutson, Ben Sheldon, Darjono & Yus Rusila Noor Summary As part of the more general studies of the Barito Ulu Project, a detailed study was made of the avifauna in July-September 1989. The survey area, which lies at the geographical centre of the island of Borneo, consists mainly of primary forest in hilly terrain, and this Is the first detailed study that has been made In the hills of Kalimantan for many decades. Aweek was also spent in montane forest at 800-1000 m. An appendix lists 226 species that were recorded. The avifauna includes 15 Bornean endemics, and extensions to known range are made for Splzaetus alboniger, Arborophila hyperythra and Megalaima eximia. Data are provided also on 20 species for which there are no recent Kalimantan records. While species described as 'slope specialists' predominated, the presence of some 26 'extreme lowland specialists' may have significance for conservation, for example Lophura erythrophthalma, Melanoperdix nigra. Pitta baudi, Malacopteron albogulare and Pityriasis gymnocephala. Ringkasan Telah dilaksanakan suatu penelitian mendalam tentang fauna burung di daerah Barito Ulu pada bulan Juli- September 1989, sebagai bagian dari proyek Barito Ulu. Ini merupakan penelitian mendalam pertama yang pernah dilakukan di perbukitan Kalimantan, untuk beberapa dekade terakhir ini. Juga dilakukan penelitian selama satu minggu di daerah pegunungan pada ketinggian 800-1000 m. Daerah penelitian terietak di tengah pulau Kalimantan yang terutama terdiri dari hutan primer perbukitan. -

Yellow Warbler Is Widespread in Southern and Eastern Africa and Is Not Included in the Lists of Threatened Species (Brooke 1984B; Yellow Warbler Collar Et Al
244 Sylviidae: warblers, apalises, crombecs, eremomelas, cisticolas and prinias southern African range is occupied by the nominate race, but birds on the Mashonaland plateau in Zimbabwe are considered subspecifically distinct (Clancey 1980b). It keeps low down in the vegetation and is easily over- looked unless singing. When disturbed it drops down into the vegetation and creeps away (Maclean 1993b). Habitat: It has a preference for scattered scrub and rank vegetation along streams and gullies, and is often recorded at the edge of evergreen forest or woodland areas surround- ing vleis, reedbeds or dams (Cyrus & Robson 1980; Ginn et al. 1989; Maclean 1993b). Its primary stronghold is in the moist vleis, rivers and wetland areas of the Eastern Zimbabwe Highlands and Miombo, where it had the high- est reporting rates, followed by several other biomes to the south. Movements: The models reveal no evidence for seasonal movements, and it appears to be resident throughout its range. Reporting rates in the eastern part of the sub- continent peak in summer, during breeding when it is sing- ing and displaying, making it more conspicuous. Although no evidence of movements can be detected in the atlas data, it apparently undergoes post-breeding altitudinal migration, moving to lower altitudes during the winter months (Cyrus & Robson 1980; Irwin 1981; Ginn et al. 1989; Johnson & Maclean 1994). Breeding: Breeding was recorded in the tropical and sub- tropical areas on the east of the subcontinent. Atlas data showed breeding activity September–April with a peak November–December, which accords with published infor- mation (Dean 1971; Irwin 1981; Tarboton et al.