Interactions with Mycorrhizal Fungi in Two Closely Related Hybridizing Orchid Species
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Impact of Climate Change on the Distribution of Four Closely Related Orchis (Orchidaceae) Species
diversity Article Impact of Climate Change on the Distribution of Four Closely Related Orchis (Orchidaceae) Species Alexandra Evans *, Sam Janssens and Hans Jacquemyn Department of Biology, Plant Conservation and Population Biology, KU Leuven, B-3001 Leuven, Belgium; [email protected] (S.J.); [email protected] (H.J.) * Correspondence: [email protected] Received: 15 July 2020; Accepted: 11 August 2020; Published: 13 August 2020 Abstract: Long-term monitoring programs and population demographic models have shown that the population dynamics of orchids are to a large extent dependent on prevailing weather conditions, suggesting that the changes in climatic conditions can have far reaching effects on the population dynamics and hence the distribution of orchids. Although a better understanding of the effects of climate change on the distribution of plants has become increasingly important during the final years, only a few studies have investigated the effects of changing temperature and precipitation on the distribution of orchids. In this study, we investigated the impact of climate change on the distribution of four terrestrial orchid species (Orchis anthropophora, Orchis militaris, Orchis purpurea and Orchis simia). Using bioclimatic data for current and future climate scenarios, habitat suitability, range shifts and the impact of different abiotic factors on the range of each species were modelled using Maxent. The results revealed an increase in suitable habitat area for O. anthropophora, O. purpurea and O. simia under each RCP (Representative Concentration Pathway) scenario, while a decrease was observed for O. militaris. Furthermore, all four of the orchids showed a shift to higher latitudes under the three RCPs leading to a significant range extension under mild climate change. -

(2008) Morphometric and Population Genetic Analyses
Botanical Journal of the Linnean Society, 2008, 157, 687–711. With 11 figures Morphometric and population genetic analyses elucidate the origin, evolutionary significance and conservation implications of Orchis ¥angusticruris (O. purpurea ¥ O. simia), a hybrid orchid new to Britain RICHARD M. BATEMAN*, RHIAN J. SMITH and MICHAEL F. FAY Jodrell Laboratory, Royal Botanic Gardens Kew, Richmond, Surrey TW9 3DS, UK Received 16 January 2008; accepted for publication 17 March 2008 We report the first confirmed occurrence in Britain of Orchis ¥ angusticruris Franch. ex Rouy, a hybrid between two closely related orchid species of anthropomorphic Orchis (O. purpurea Huds. ¥ O. simia Lam.) that hybridize frequently in Continental Europe. Seven individual hybrids, most likely F1 plants representing a single interspe- cific pollination event, first flowered with both parents in May 2006 at a nature reserve in the Chiltern Hills near Goring, Oxfordshire. Univariate and multivariate morphometric analyses (43 characters plus 12 indices), internal transcribed spacer sequencing, plastid microsatellites and amplified fragment length polymorphism (AFLP) analyses together readily separate the parents and confirm that O. purpurea was the ovule parent and O. simia the pollen parent, presumably reflecting the greater frequency and/or later flowering period of the latter at the site. This study reinforces a more general observation that, in most orchids, the ovule parent contributes substantially more to the hybrid phenotype than does the pollen parent, perhaps reflecting cytoplasmic inheritance. In contrast, the hybrids are placed closer to O. simia than to O. purpurea in the AFLP tree. Apparently recent arrivals, the few O. purpurea plants at Goring contrast genetically with the two other small populations of this species known in the Chilterns, but rather are consistent with relatively uncommon Continental populations. -

Girosnotizie 15
GIROS Notizie n. 15 - 2000 GIROS NOTIZIE G.I.R.O.S. Notiziario per i Soci Gruppo Italiano per la Ricerca sulle o Orchidee Spontanee Anno 2000 - N 15 web: http://astrpi.difi.unipi.it/Orchids/Giros.html http://astrpi.difi.unipi.it/Orchids-NEW Redazione e impaginazione a cura di: e-mail: [email protected] Bruno Barsella [email protected] ([email protected]) Mauro Biagioli ([email protected]) Sede legale: Paolo Grünanger ([email protected]) Via Testi, 7 - 48018 FAENZA (RA) Giuliano Pacifico Tel# 0546/30833 (Paolo Liverani) ([email protected]) Comitato Scientifico: Segreteria: Carlo Del Prete ([email protected]) Via Rosi, 21 - 55100 LUCCA (LU) Paolo Grünanger ([email protected]) Tel# 0583/492169 (Marcello Pieruccini) Giorgio Perazza Quota sociale 2000: L. 50.000 Grafica copertine: o Patrizia Cini e da versare sul c.c.p. n 13552559 intestato a: Bruno Barsella Gruppo Micologico M. Danesi A.M.B. 55029 - Ponte a Moriano - Lucca Sulla copertina: Cariche sociali per il triennio 2000-2002 Epipogium aphyllum Swartz foto di Rolando Romolini Consiglio Direttivo: Paolo Liverani (Presidente) Bruno Barsella (Vicepresidente) Marcello Pieruccini (Segretario) Stivi Betti (Tesoriere) M. Elisabetta Aloisi Masella NOTA DELLA REDAZIONE: Mauro Biagioli Rolando Romolini Ringraziamo i numerosi soci che hanno contribuito alla realizzazione di questo Sindaci Revisori: n u m e ro di “GIROS Notizie”. Rinnoviamo l’invito a collaborare alla Fulvio Fiesoli stesura dei notiziari inviando alla Claudio Merlini (Coordinatore) Redazione articoli, fotografie e suggeri - Michele Petroni menti. Nella ultima pagina di questo numero si vedano le norme redazionali da seguire per sottoporre manoscritti, disegni e immagini. -

Ornamental Plants in Different Approaches
Ornamental Plants in Different Approaches Assoc. Prof. Dr. Arzu ÇIĞ cultivation sustainibility ecology propagation ORNAMENTAL PLANTS IN DIFFERENT APPROACHES EDITOR Assoc. Prof. Dr. Arzu ÇIĞ AUTHORS Atilla DURSUN Feran AŞUR Husrev MENNAN Görkem ÖRÜK Kazım MAVİ İbrahim ÇELİK Murat Ertuğrul YAZGAN Muhemet Zeki KARİPÇİN Mustafa Ercan ÖZZAMBAK Funda ANKAYA Ramazan MAMMADOV Emrah ZEYBEKOĞLU Şevket ALP Halit KARAGÖZ Arzu ÇIĞ Jovana OSTOJIĆ Bihter Çolak ESETLILI Meltem Yağmur WALLACE Elif BOZDOGAN SERT Murat TURAN Elif AKPINAR KÜLEKÇİ Samim KAYIKÇI Firat PALA Zehra Tugba GUZEL Mirjana LJUBOJEVIĆ Fulya UZUNOĞLU Nazire MİKAİL Selin TEMİZEL Slavica VUKOVIĆ Meral DOĞAN Ali SALMAN İbrahim Halil HATİPOĞLU Dragana ŠUNJKA İsmail Hakkı ÜRÜN Fazilet PARLAKOVA KARAGÖZ Atakan PİRLİ Nihan BAŞ ZEYBEKOĞLU M. Anıl ÖRÜK Copyright © 2020 by iksad publishing house All rights reserved. No part of this publication may be reproduced, distributed or transmitted in any form or by any means, including photocopying, recording or other electronic or mechanical methods, without the prior written permission of the publisher, except in the case of brief quotations embodied in critical reviews and certain other noncommercial uses permitted by copyright law. Institution of Economic Development and Social Researches Publications® (The Licence Number of Publicator: 2014/31220) TURKEY TR: +90 342 606 06 75 USA: +1 631 685 0 853 E mail: [email protected] www.iksadyayinevi.com It is responsibility of the author to abide by the publishing ethics rules. Iksad Publications – 2020© ISBN: 978-625-7687-07-2 Cover Design: İbrahim KAYA December / 2020 Ankara / Turkey Size = 16 x 24 cm CONTENTS PREFACE Assoc. Prof. Dr. Arzu ÇIĞ……………………………………………1 CHAPTER 1 DOUBLE FLOWER TRAIT IN ORNAMENTAL PLANTS: FROM HISTORICAL PERSPECTIVE TO MOLECULAR MECHANISMS Prof. -

Pdf of JHOS July 2007
JJoouurrnnaall of the HHAARRDDYY OORRCCHHIIDD SSOOCCIIEETTYY Vol. 4 No. 3 (45) July 2007 JOURNAL of the HARDY ORCHID SOCIETY Vol. 4 No. 3 (45) July 2007 The Hardy Orchid Society Our aim is to promote interest in the study of Native European Orchids and those from similar temperate climates throughout the world. We cover such varied aspects as field study, cultivation and propagation, photography, taxonomy and systematics, and practical conservation. We welcome articles relating to any of these subjects, which will be considered for publication by the editorial committee. Please send your submissions to the Editor, and please structure your text according to the “Advice to Authors ” (see website, January 2004 Journal or contact the Editor). The Hardy Orchid Society Committee President: Prof. Richard Bateman, Jodrell Laboratory, Royal Botanic Gardens Kew, Richmond, Surrey, TW9 3DS Chairman: Tony Hughes, 8 Birchwood Road, Malvern, Worcs., WR14 1LD, [email protected] Vice-Chairman & Field Meeting Co-ordinator: David Hughes, Linmoor Cottage, Highwood, Ringwood, Hants., BH24 3LE, [email protected] Secretary: Richard Manuel, Wye View Cottage, Leys Hill, Ross-on-Wye, Herefordshire, HR9 5QU, [email protected] Treasurer: Iain Wright, The Windmill, Vennington, Westbury, Shewsbury, Shropshire, SY5 9RG, [email protected] Membership Secretary: Celia Wright, The Windmill, Vennington, Westbury, Shewsbury, Shropshire, SY5 9RG, [email protected] Show Secretary: Eric Webster, 25 Highfields Drive, Loughborough, Leics., LE11 -

Greece (Orchids)
Orchids Among the Thorns, or BY SPIRO KASOMENAKIS/PHOTOGRAPHS, UNLESS OTHERWISE CREDITED, BY THE AUTHOR A group of Orchis italica in typical phrygana habitat. The insert is a closeup of a single infl orescence. Orchids of Crete and Attica GREECE, A SMALL country on the southeast end of Europe, is well known for its spring fl ora, especially to orchid lovers, as it harbors about 227 species of orchids, of which 74 are endemic. Granted, for the general public that fl ocks to Greece and its islands for the wonderful beaches, orchids may not be the fi rst thing that comes to mind, but the devoti on these plants (especially the genus Ophrys) seem to inspire, in both botanical and hobbyist circles, is worth a closer look. Our trip was organized by the Orchid Conservati on Alliance (OCA), a nonprofi t conservati on organizati on that regularly takes members on orchid-related trips. We concentrated on western Crete, beginning our odyssey at Chania, and later moving our base to the seaside village of Plakias, with a few days at the end of the trip on the mainland to see some archaeological sites and more orchids! Crete is a microcosm in itself, being a large and self-sufficient island on the southernmost part of Greece. The landscape is dominated by the snow- capped White Mountains, visible from the city of Chania. Bound by the Aegean Sea on its northern shores, and the Libyan Sea on its southern, its flora shows the influences of both east and west, north and south! Fifteen of the 70 or so species of orchids on the island are found nowhere else. -

The Effects of Inflorescence Size and Flower Position on Female Reproductive Success in Three Deceptive Orchids
Botanical Studies (2010) 51: 351-356. REProductiVE BIOLOGY The effects of inflorescence size and flower position on female reproductive success in three deceptive orchids Giuseppe PELLEGRINO*, Francesca BELLUSCI, and Aldo MUSACCHIO Dipartimento di Ecologia, Università della Calabria, via P. Bucci 6/B, I-87036 Arcavacata di Rende, Cosenza, Italy (Received January 8, 2009; Accepted January 21, 2010) ABSTRACT. Most Mediterranean orchids are deceptive, offering no reward to insect pollinators. In the absence of a reward, inflorescence size and flower position within inflorescence may be associated with the timing and the numbers of pollinators attracted, thus influencing the final reproductive success. Pollination biology and breeding system were investigated in three deceptive orchids, Orchis italica, O. anthropophora, and Anacamptis papilionacea. Although all examined orchids were self-compatible, bagged inflorescences produced no fruits. Artificial pollination resulted in a 76-79% fruit set by induced autogamy, a 70-76% one by geitonogamous pollination, and a 79-87% one by allogamous pollination. The natural fruit set in the open- pollinated control was 14-16%. Fruit production was neither related to the inflorescence size, nor to the number and position of flowers within the flowering spike, suggesting that variation in these floral traits does not influence pollinator attraction or female reproductive success. Keywords: Anacamptis; Breeding system; Deceptive orchid; Hand-pollination; Orchis; Reproductive success. INTRODUCTION Pollinator abundance and resource limitation are factors which may limit reproductive success in orchids (Zim- The Orchidaceae is one of the largest families of merman and Aide, 1989). Inflorescence size in such cases flowering plants (up to 35,000 species), making up nearly should represent a trade off between the need to attract 10% of all flowering plant species in the world (Dressler, scarce pollinators and the quantity of resources available 1993). -

Dottorato Di Ricerca
Università degli Studi di Cagliari DOTTORATO DI RICERCA IN SCIENZE E TECNOLOGIE DELLA TERRA E DELL'AMBIENTE Ciclo XXXI Patterns of reproductive isolation in Sardinian orchids of the subtribe Orchidinae Settore scientifico disciplinare di afferenza Botanica ambientale e applicata, BIO/03 Presentata da: Dott. Michele Lussu Coordinatore Dottorato Prof. Aldo Muntoni Tutor Dott.ssa Michela Marignani Co-tutor Prof.ssa Annalena Cogoni Dott. Pierluigi Cortis Esame finale anno accademico 2018 – 2019 Tesi discussa nella sessione d’esame Febbraio –Aprile 2019 2 Table of contents Chapter 1 Abstract Riassunto………………………………………………………………………………………….. 4 Preface ………………………………………………………………………………………………………. 6 Chapter 2 Introduction …………………………………………………………………………………………………. 8 Aim of the study…………………………………………………………………………………………….. 14 Chapter 3 What we didn‘t know, we know and why is important working on island's orchids. A synopsis of Sardinian studies……………………………………………………………………………………………………….. 17 Chapter 4 Ophrys annae and Ophrys chestermanii: an impossible love between two orchid sister species…………. 111 Chapter 5 Does size really matter? A comparative study on floral traits in two different orchid's pollination strategies……………………………………………………………………………………………………. 133 Chapter 6 General conclusions………………………………………………………………………………………... 156 3 Chapter 1 Abstract Orchids are globally well known for their highly specialized mechanisms of pollination as a result of their complex biology. Based on natural selection, mutation and genetic drift, speciation occurs simultaneously in organisms linking them in complexes webs called ecosystems. Clarify what a species is, it is the first step to understand the biology of orchids and start protection actions especially in a fast changing world due to human impact such as habitats fragmentation and climate changes. I use the biological species concept (BSC) to investigate the presence and eventually the strength of mechanisms that limit the gene flow between close related taxa. -

Wildlife Travel Cyprus Spring 2015
Cyprus trip report, 11th to 20th March 2015 WILDLIFE TRAVEL Cyprus 2015 Cyprus trip report, 11th to 20th March 2015 Cover: Ophrys cinereophila www.wildlife-travel.co.uk 2 Cyprus trip report, 11th to 20th March 2015 11th March. Arrival at Paphos airport and after meeting Yiannis transfer to our welcoming hotel in the Akamas peninsula. A short walk in the late afternoon (shortly after arriving at the hotel), set the scene for much of the birding that was to come. The hotel gardens and neighbouring fields held many familiar species found commonly in the UK including Swallow, House Martin, Collared Dove, House Sparrow, Great Tit, Chiffchaff, Blackcap, Corn Bunting, Chaffinch, Greenfinch, Hooded Crow and Jackdaw for example. All these species were widespread and seen on almost every day of our trip. Of more interest were the Hawfinches seen by the chicken coop across the road (the species winters in Cyprus), the Serins heard twittering nearby, and the first of many Sardinian Warblers and Zitting Cisticolas. A distant singing Black Francolin hinted that despite recent housing developments we would get to see this strikingly beautiful but often elusive game bird that, in Europe, breeds only in Cyprus. We also heard the second of Cyprus's game birds, the Chukar – a bird very similar to the better known Red-legged Partridge. 12th March. After breakfast we set off on our first excursion towards the Baths of Aphrodite. Many interesting plants were found by the roadside, including Prasium majus, Carob, Gladiolus italicus, Bellevalia trifoliata and our first orchids; Ophrys flavomarginata and Ophrys sicula. -

Download Download
Dipartimento di Biologia Ambientale ANNALI DI BOTANICA COENOLOGY AND PLANT ECOLOGY ANNALI DI BOTANICA Volume 7, 2017 V o l u m e 7 Ann. Bot. (Roma), 2017, 7: 71–74 Published in Rome (Italy) 2 ISSN 0365-0812 0 1 7 Journal homepage: http://annalidibotanica.uniroma1.it Pietro Romualdo Pirotta, founder, 1884 NOTES ORCHIS ITALICA POIR. (ORCHIDACEAE): REDISCOVERY AFTER FOUR CENTURIES OF A PRESUMABLY EXTINCT SPECIES IN MT. VESUVIUS, ITALY STINCA A.1,2 * 1 Department of Agriculture, University of Naples Federico II, via Università 100, 80055 Portici (Naples), Italy 2 Department of Environmental Biological and Pharmaceutical Sciences and Technologies, University of Campania Luigi Vanvitelli, via Vivaldi 43, 81100, Caserta, Italy *Corresponding author: Telephone: 0812539369; e-mail: [email protected] (RECEIVED 26 MAY 2016; RECEIVED IN REVISED FORM 24 NOVEMBER 2016; ACCEPTED 25 NOVEMBER 2016) ABSTRACT – Orchis italica Poir. (Orchidaceae), thought to be extinct on Mt. Vesuvius, has been rediscovered after a gap of four centuries. Its ecology and conservation status are discussed, and photographs provided. KEYWORDS: BIODIVERSITY, CONSERVATION, FIELD RESEARCH, ORCHIDS, VASCULAR FLORA INTRODUCTION The Orchidaceae comprise approximately 850 genera and inflorescence is a spike, conical at first and becoming ovoid, 20,000 species (Dressler, 1993). As many orchids are threatened with up to 40 zygomorphic and epigynous flowers; 6 perianth with extinction, whether local, regional or global, to promote segments in 2 whorls, lanceolate, acuminate, convergent into their conservation the whole family has been inserted in a galea, pale pink with purple veins; pink labellum 12-20 mm, the Appendices of the Convention on International Trade longer than wide, directed downwards owing to the ovary in Endangered Species of Wild Fauna and Flora (CITES). -

Download Download
Firenze University Press Caryologia www.fupress.com/caryologia International Journal of Cytology, Cytosystematics and Cytogenetics Comparison of the Evolution of Orchids with that of Bats Citation: A. Lima-de-Faria (2020) Comparison of the Evolution of Orchids with that of Bats. Caryologia 73(2): 51-61. doi: 10.13128/caryologia-891 Antonio Lima-de-Faria Received: February 12, 2020 Professor Emeritus of Molecular Cytogenetics, Lund University, Lund, Sweden E-mail: [email protected] Accepted: April 16, 2020 Published: July 31, 2020 Abstract. The evolution of orchids and bats is an example of DNA’s own evolution which has resulted in structures and functions which are not necessarily related to any Copyright: © 2020 A. Lima-de-Faria. obvious advantage to the organism. The flowers of orchids resemble: humans, apes, liz- This is an open access, peer-reviewed article published by Firenze University ards, frogs and even shoes. The faces of bats resemble plant leaves but also horseshoes. Press (http://www.fupress.com/caryo- These similarities are not accidental because they emerge repeatedly in different gen- logia) and distributed under the terms era and different families. This evolutionary situation bewildered botanists and zoolo- of the Creative Commons Attribution gists for many years, but is now elucidated by the molecular unification of plants and License, which permits unrestricted animals derived from the following evidence: (1) Contrary to expectation, plant and use, distribution, and reproduction animal cells (including those of humans) could be fused and the human chromosomes in any medium, provided the original were seen dividing in the plant cytoplasm. (2) Orchids, bats and humans have about author and source are credited. -
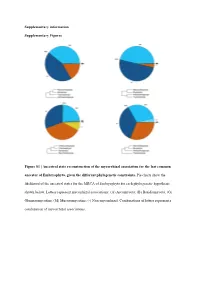
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea